William Ratliff
Development and Validation of ML-DQA -- a Machine Learning Data Quality Assurance Framework for Healthcare
Aug 04, 2022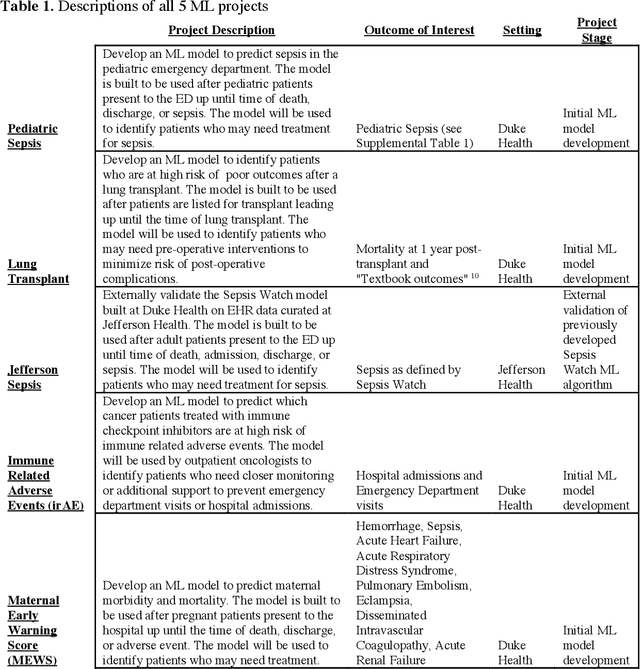
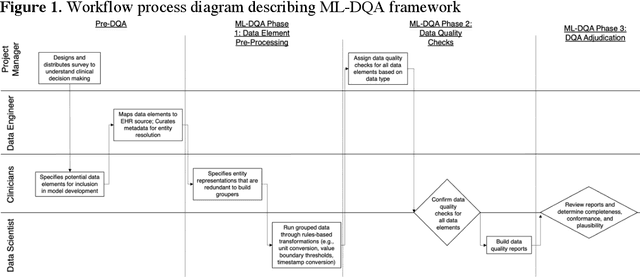
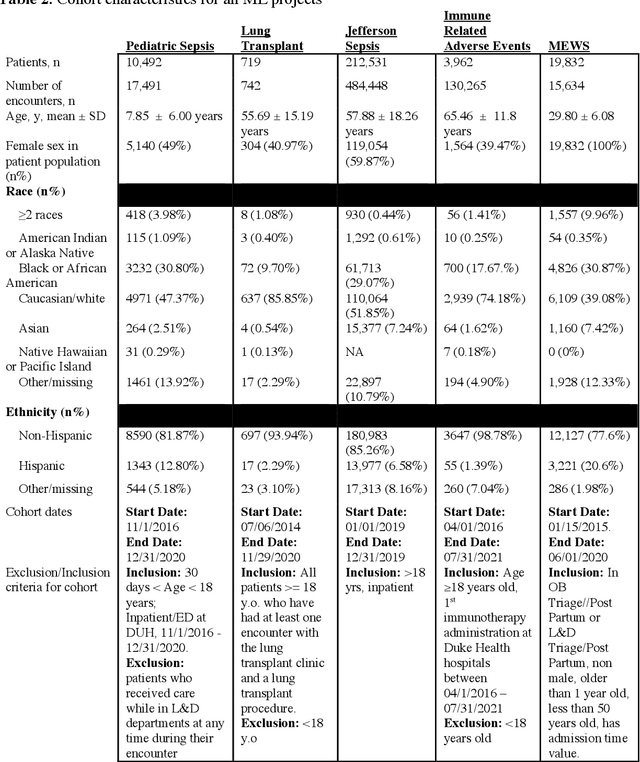
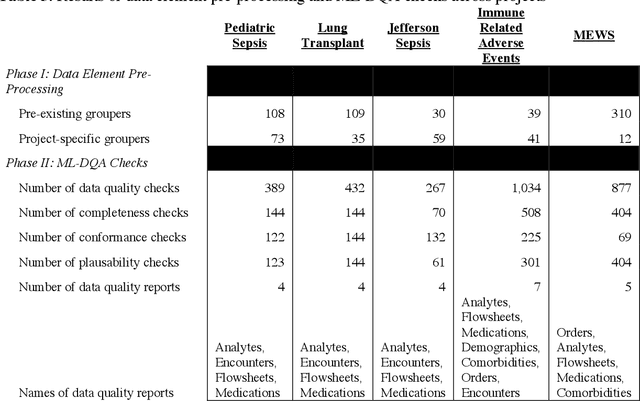
Abstract:The approaches by which the machine learning and clinical research communities utilize real world data (RWD), including data captured in the electronic health record (EHR), vary dramatically. While clinical researchers cautiously use RWD for clinical investigations, ML for healthcare teams consume public datasets with minimal scrutiny to develop new algorithms. This study bridges this gap by developing and validating ML-DQA, a data quality assurance framework grounded in RWD best practices. The ML-DQA framework is applied to five ML projects across two geographies, different medical conditions, and different cohorts. A total of 2,999 quality checks and 24 quality reports were generated on RWD gathered on 247,536 patients across the five projects. Five generalizable practices emerge: all projects used a similar method to group redundant data element representations; all projects used automated utilities to build diagnosis and medication data elements; all projects used a common library of rules-based transformations; all projects used a unified approach to assign data quality checks to data elements; and all projects used a similar approach to clinical adjudication. An average of 5.8 individuals, including clinicians, data scientists, and trainees, were involved in implementing ML-DQA for each project and an average of 23.4 data elements per project were either transformed or removed in response to ML-DQA. This study demonstrates the importance role of ML-DQA in healthcare projects and provides teams a framework to conduct these essential activities.
Malignancy Prediction and Lesion Identification from Clinical Dermatological Images
Apr 02, 2021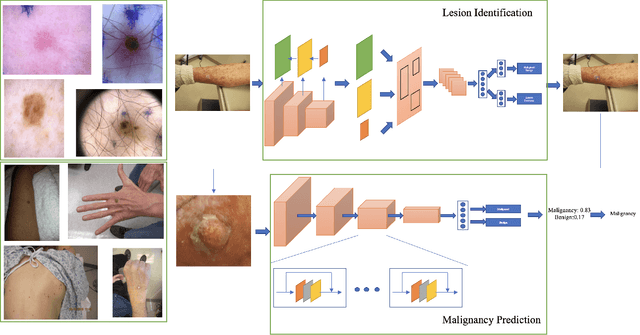
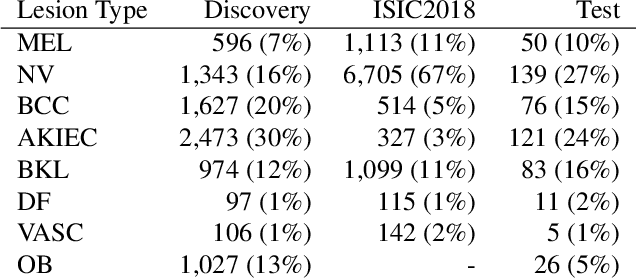
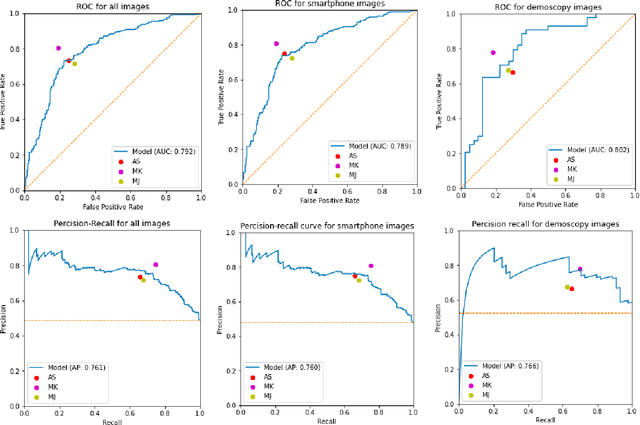
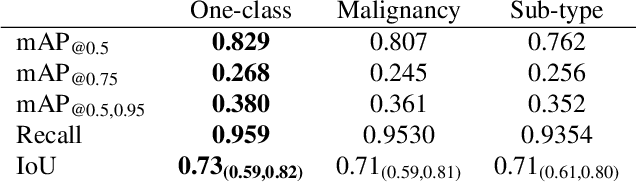
Abstract:We consider machine-learning-based malignancy prediction and lesion identification from clinical dermatological images, which can be indistinctly acquired via smartphone or dermoscopy capture. Additionally, we do not assume that images contain single lesions, thus the framework supports both focal or wide-field images. Specifically, we propose a two-stage approach in which we first identify all lesions present in the image regardless of sub-type or likelihood of malignancy, then it estimates their likelihood of malignancy, and through aggregation, it also generates an image-level likelihood of malignancy that can be used for high-level screening processes. Further, we consider augmenting the proposed approach with clinical covariates (from electronic health records) and publicly available data (the ISIC dataset). Comprehensive experiments validated on an independent test dataset demonstrate that i) the proposed approach outperforms alternative model architectures; ii) the model based on images outperforms a pure clinical model by a large margin, and the combination of images and clinical data does not significantly improves over the image-only model; and iii) the proposed framework offers comparable performance in terms of malignancy classification relative to three board certified dermatologists with different levels of experience.
"The Human Body is a Black Box": Supporting Clinical Decision-Making with Deep Learning
Dec 07, 2019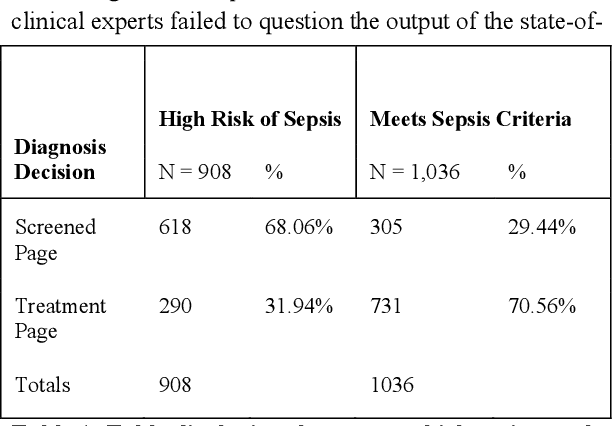
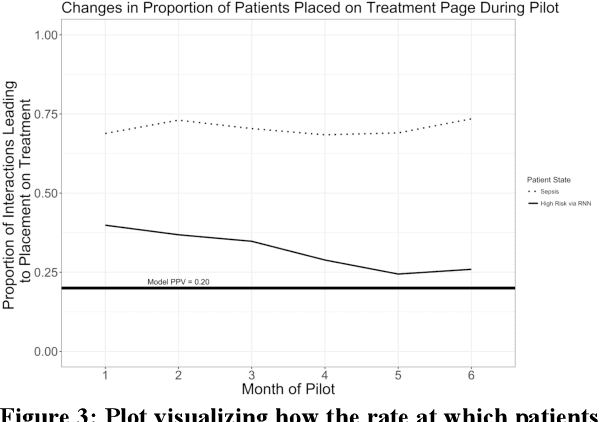
Abstract:Machine learning technologies are increasingly developed for use in healthcare. While research communities have focused on creating state-of-the-art models, there has been less focus on real world implementation and the associated challenges to accuracy, fairness, accountability, and transparency that come from actual, situated use. Serious questions remain under examined regarding how to ethically build models, interpret and explain model output, recognize and account for biases, and minimize disruptions to professional expertise and work cultures. We address this gap in the literature and provide a detailed case study covering the development, implementation, and evaluation of Sepsis Watch, a machine learning-driven tool that assists hospital clinicians in the early diagnosis and treatment of sepsis. We, the team that developed and evaluated the tool, discuss our conceptualization of the tool not as a model deployed in the world but instead as a socio-technical system requiring integration into existing social and professional contexts. Rather than focusing on model interpretability to ensure a fair and accountable machine learning, we point toward four key values and practices that should be considered when developing machine learning to support clinical decision-making: rigorously define the problem in context, build relationships with stakeholders, respect professional discretion, and create ongoing feedback loops with stakeholders. Our work has significant implications for future research regarding mechanisms of institutional accountability and considerations for designing machine learning systems. Our work underscores the limits of model interpretability as a solution to ensure transparency, accuracy, and accountability in practice. Instead, our work demonstrates other means and goals to achieve FATML values in design and in practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge