Christine Park
Improving Image Classification of Knee Radiographs: An Automated Image Labeling Approach
Sep 06, 2023Abstract:Large numbers of radiographic images are available in knee radiology practices which could be used for training of deep learning models for diagnosis of knee abnormalities. However, those images do not typically contain readily available labels due to limitations of human annotations. The purpose of our study was to develop an automated labeling approach that improves the image classification model to distinguish normal knee images from those with abnormalities or prior arthroplasty. The automated labeler was trained on a small set of labeled data to automatically label a much larger set of unlabeled data, further improving the image classification performance for knee radiographic diagnosis. We developed our approach using 7,382 patients and validated it on a separate set of 637 patients. The final image classification model, trained using both manually labeled and pseudo-labeled data, had the higher weighted average AUC (WAUC: 0.903) value and higher AUC-ROC values among all classes (normal AUC-ROC: 0.894; abnormal AUC-ROC: 0.896, arthroplasty AUC-ROC: 0.990) compared to the baseline model (WAUC=0.857; normal AUC-ROC: 0.842; abnormal AUC-ROC: 0.848, arthroplasty AUC-ROC: 0.987), trained using only manually labeled data. DeLong tests show that the improvement is significant on normal (p-value<0.002) and abnormal (p-value<0.001) images. Our findings demonstrated that the proposed automated labeling approach significantly improves the performance of image classification for radiographic knee diagnosis, allowing for facilitating patient care and curation of large knee datasets.
Automated Grading of Radiographic Knee Osteoarthritis Severity Combined with Joint Space Narrowing
Mar 16, 2022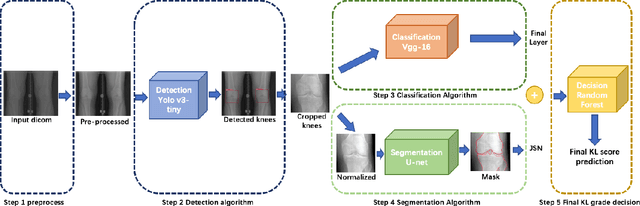
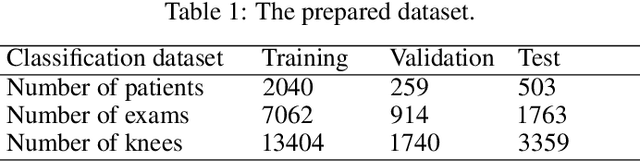
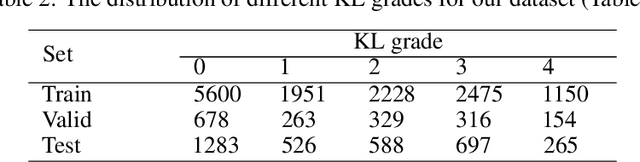
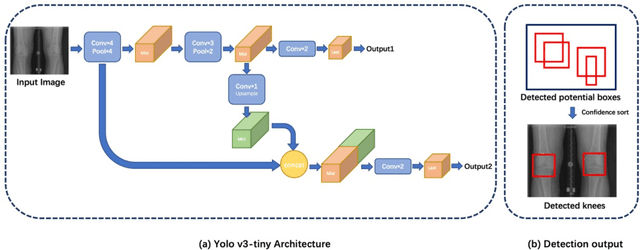
Abstract:The assessment of knee osteoarthritis (KOA) severity on knee X-rays is a central criteria for the use of total knee arthroplasty. However, this assessment suffers from imprecise standards and a remarkably high inter-reader variability. An algorithmic, automated assessment of KOA severity could improve overall outcomes of knee replacement procedures by increasing the appropriateness of its use. We propose a novel deep learning-based five-step algorithm to automatically grade KOA from posterior-anterior (PA) views of radiographs: (1) image preprocessing (2) localization of knees joints in the image using the YOLO v3-Tiny model, (3) initial assessment of the severity of osteoarthritis using a convolutional neural network-based classifier, (4) segmentation of the joints and calculation of the joint space narrowing (JSN), and (5), a combination of the JSN and the initial assessment to determine a final Kellgren-Lawrence (KL) score. Furthermore, by displaying the segmentation masks used to make the assessment, our algorithm demonstrates a higher degree of transparency compared to typical "black box" deep learning classifiers. We perform a comprehensive evaluation using two public datasets and one dataset from our institution, and show that our algorithm reaches state-of-the art performance. Moreover, we also collected ratings from multiple radiologists at our institution and showed that our algorithm performs at the radiologist level. The software has been made publicly available at https://github.com/MaciejMazurowski/osteoarthritis-classification.
Malignancy Prediction and Lesion Identification from Clinical Dermatological Images
Apr 02, 2021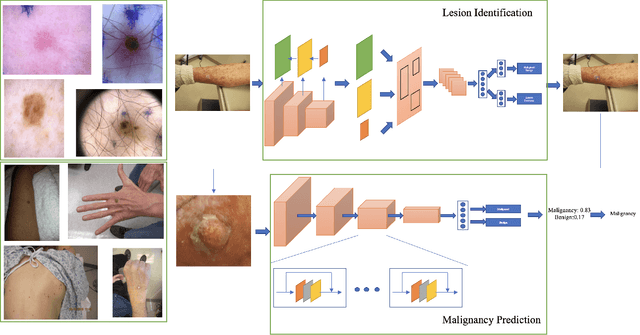
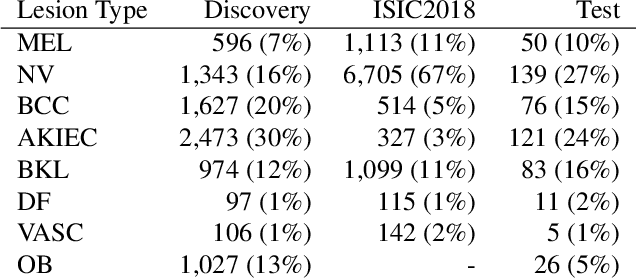
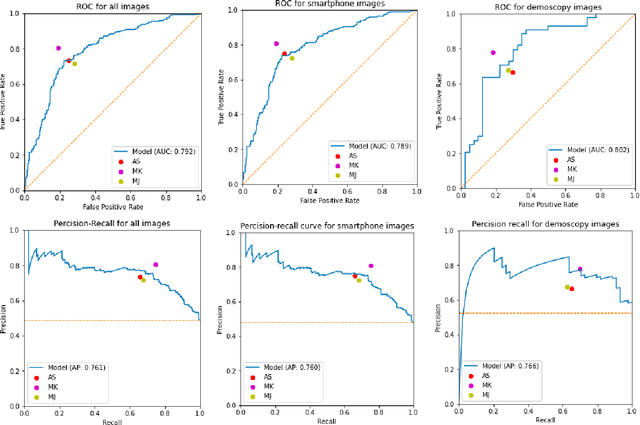
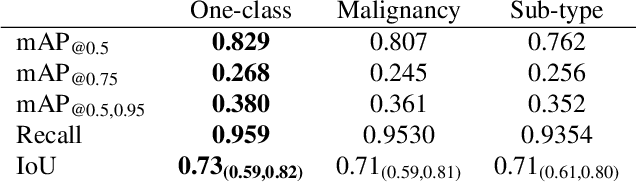
Abstract:We consider machine-learning-based malignancy prediction and lesion identification from clinical dermatological images, which can be indistinctly acquired via smartphone or dermoscopy capture. Additionally, we do not assume that images contain single lesions, thus the framework supports both focal or wide-field images. Specifically, we propose a two-stage approach in which we first identify all lesions present in the image regardless of sub-type or likelihood of malignancy, then it estimates their likelihood of malignancy, and through aggregation, it also generates an image-level likelihood of malignancy that can be used for high-level screening processes. Further, we consider augmenting the proposed approach with clinical covariates (from electronic health records) and publicly available data (the ISIC dataset). Comprehensive experiments validated on an independent test dataset demonstrate that i) the proposed approach outperforms alternative model architectures; ii) the model based on images outperforms a pure clinical model by a large margin, and the combination of images and clinical data does not significantly improves over the image-only model; and iii) the proposed framework offers comparable performance in terms of malignancy classification relative to three board certified dermatologists with different levels of experience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge