Maciej Mazurowski
Language Models and Retrieval Augmented Generation for Automated Structured Data Extraction from Diagnostic Reports
Sep 18, 2024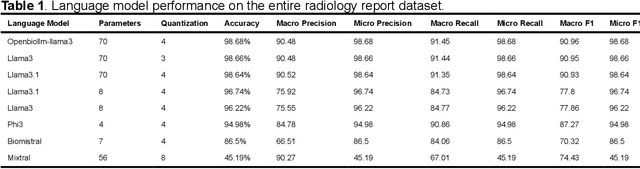

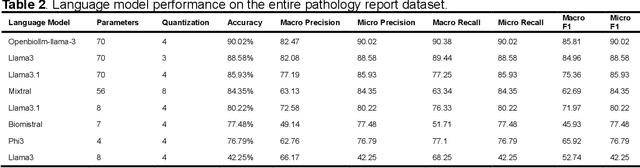

Abstract:Purpose: To develop and evaluate an automated system for extracting structured clinical information from unstructured radiology and pathology reports using open-weights large language models (LMs) and retrieval augmented generation (RAG), and to assess the effects of model configuration variables on extraction performance. Methods and Materials: The study utilized two datasets: 7,294 radiology reports annotated for Brain Tumor Reporting and Data System (BT-RADS) scores and 2,154 pathology reports annotated for isocitrate dehydrogenase (IDH) mutation status. An automated pipeline was developed to benchmark the performance of various LMs and RAG configurations. The impact of model size, quantization, prompting strategies, output formatting, and inference parameters was systematically evaluated. Results: The best performing models achieved over 98% accuracy in extracting BT-RADS scores from radiology reports and over 90% for IDH mutation status extraction from pathology reports. The top model being medical fine-tuned llama3. Larger, newer, and domain fine-tuned models consistently outperformed older and smaller models. Model quantization had minimal impact on performance. Few-shot prompting significantly improved accuracy. RAG improved performance for complex pathology reports but not for shorter radiology reports. Conclusions: Open LMs demonstrate significant potential for automated extraction of structured clinical data from unstructured clinical reports with local privacy-preserving application. Careful model selection, prompt engineering, and semi-automated optimization using annotated data are critical for optimal performance. These approaches could be reliable enough for practical use in research workflows, highlighting the potential for human-machine collaboration in healthcare data extraction.
Improving Image Classification of Knee Radiographs: An Automated Image Labeling Approach
Sep 06, 2023Abstract:Large numbers of radiographic images are available in knee radiology practices which could be used for training of deep learning models for diagnosis of knee abnormalities. However, those images do not typically contain readily available labels due to limitations of human annotations. The purpose of our study was to develop an automated labeling approach that improves the image classification model to distinguish normal knee images from those with abnormalities or prior arthroplasty. The automated labeler was trained on a small set of labeled data to automatically label a much larger set of unlabeled data, further improving the image classification performance for knee radiographic diagnosis. We developed our approach using 7,382 patients and validated it on a separate set of 637 patients. The final image classification model, trained using both manually labeled and pseudo-labeled data, had the higher weighted average AUC (WAUC: 0.903) value and higher AUC-ROC values among all classes (normal AUC-ROC: 0.894; abnormal AUC-ROC: 0.896, arthroplasty AUC-ROC: 0.990) compared to the baseline model (WAUC=0.857; normal AUC-ROC: 0.842; abnormal AUC-ROC: 0.848, arthroplasty AUC-ROC: 0.987), trained using only manually labeled data. DeLong tests show that the improvement is significant on normal (p-value<0.002) and abnormal (p-value<0.001) images. Our findings demonstrated that the proposed automated labeling approach significantly improves the performance of image classification for radiographic knee diagnosis, allowing for facilitating patient care and curation of large knee datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge