Language Models and Retrieval Augmented Generation for Automated Structured Data Extraction from Diagnostic Reports
Paper and Code
Sep 18, 2024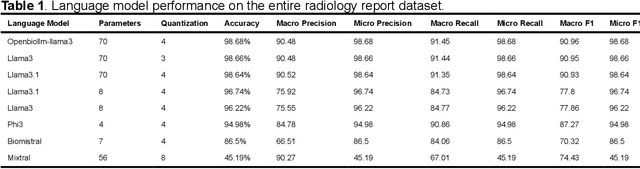

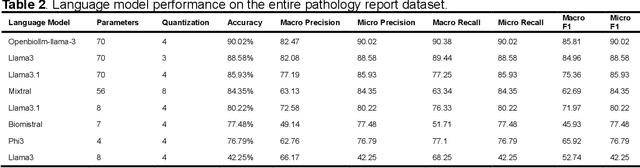

Purpose: To develop and evaluate an automated system for extracting structured clinical information from unstructured radiology and pathology reports using open-weights large language models (LMs) and retrieval augmented generation (RAG), and to assess the effects of model configuration variables on extraction performance. Methods and Materials: The study utilized two datasets: 7,294 radiology reports annotated for Brain Tumor Reporting and Data System (BT-RADS) scores and 2,154 pathology reports annotated for isocitrate dehydrogenase (IDH) mutation status. An automated pipeline was developed to benchmark the performance of various LMs and RAG configurations. The impact of model size, quantization, prompting strategies, output formatting, and inference parameters was systematically evaluated. Results: The best performing models achieved over 98% accuracy in extracting BT-RADS scores from radiology reports and over 90% for IDH mutation status extraction from pathology reports. The top model being medical fine-tuned llama3. Larger, newer, and domain fine-tuned models consistently outperformed older and smaller models. Model quantization had minimal impact on performance. Few-shot prompting significantly improved accuracy. RAG improved performance for complex pathology reports but not for shorter radiology reports. Conclusions: Open LMs demonstrate significant potential for automated extraction of structured clinical data from unstructured clinical reports with local privacy-preserving application. Careful model selection, prompt engineering, and semi-automated optimization using annotated data are critical for optimal performance. These approaches could be reliable enough for practical use in research workflows, highlighting the potential for human-machine collaboration in healthcare data extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge