Ayush Jain
Department of Computer Science and Engineering, Indian Institute of Technology Hyderabad, India
Structure Enables Effective Self-Localization of Errors in LLMs
Feb 02, 2026Abstract:Self-correction in language models remains elusive. In this work, we explore whether language models can explicitly localize errors in incorrect reasoning, as a path toward building AI systems that can effectively correct themselves. We introduce a prompting method that structures reasoning as discrete, semantically coherent thought steps, and show that models are able to reliably localize errors within this structure, while failing to do so in conventional, unstructured chain-of-thought reasoning. Motivated by how the human brain monitors errors at discrete decision points and resamples alternatives, we introduce Iterative Correction Sampling of Thoughts (Thought-ICS), a self-correction framework. Thought-ICS iteratively prompts the model to generate reasoning one discrete and complete thought at a time--where each thought represents a deliberate decision by the model--creating natural boundaries for precise error localization. Upon verification, the model localizes the first erroneous step, and the system backtracks to generate alternative reasoning from the last correct point. When asked to correct reasoning verified as incorrect by an oracle, Thought-ICS achieves 20-40% self-correction lift. In a completely autonomous setting without external verification, it outperforms contemporary self-correction baselines.
Explainable AI in Big Data Fraud Detection
Dec 17, 2025Abstract:Big Data has become central to modern applications in finance, insurance, and cybersecurity, enabling machine learning systems to perform large-scale risk assessments and fraud detection. However, the increasing dependence on automated analytics introduces important concerns about transparency, regulatory compliance, and trust. This paper examines how explainable artificial intelligence (XAI) can be integrated into Big Data analytics pipelines for fraud detection and risk management. We review key Big Data characteristics and survey major analytical tools, including distributed storage systems, streaming platforms, and advanced fraud detection models such as anomaly detectors, graph-based approaches, and ensemble classifiers. We also present a structured review of widely used XAI methods, including LIME, SHAP, counterfactual explanations, and attention mechanisms, and analyze their strengths and limitations when deployed at scale. Based on these findings, we identify key research gaps related to scalability, real-time processing, and explainability for graph and temporal models. To address these challenges, we outline a conceptual framework that integrates scalable Big Data infrastructure with context-aware explanation mechanisms and human feedback. The paper concludes with open research directions in scalable XAI, privacy-aware explanations, and standardized evaluation methods for explainable fraud detection systems.
Verification-Guided Context Optimization for Tool Calling via Hierarchical LLMs-as-Editors
Dec 15, 2025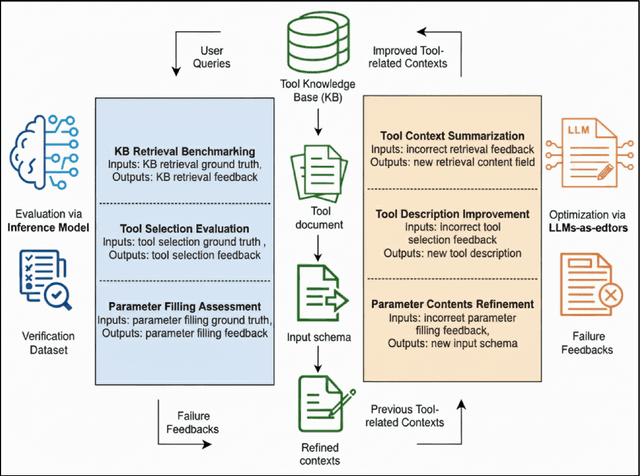
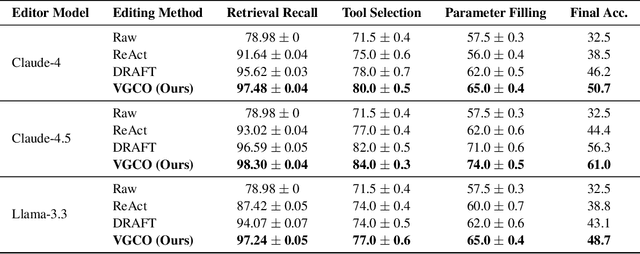
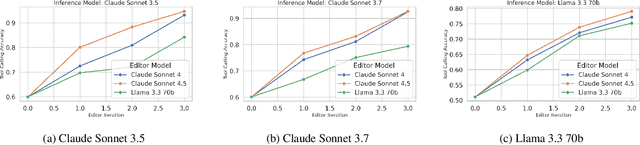
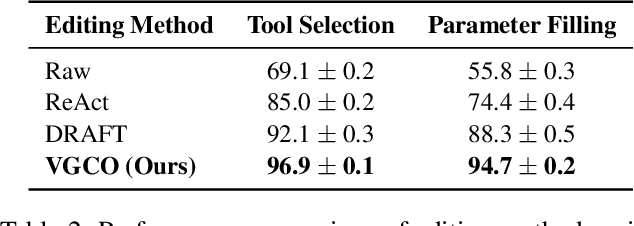
Abstract:Tool calling enables large language models (LLMs) to interact with external environments through tool invocation, providing a practical way to overcome the limitations of pretraining. However, the effectiveness of tool use depends heavily on the quality of the associated documentation and knowledge base context. These materials are usually written for human users and are often misaligned with how LLMs interpret information. This problem is even more pronounced in industrial settings, where hundreds of tools with overlapping functionality create challenges in scalability, variability, and ambiguity. We propose Verification-Guided Context Optimization (VGCO), a framework that uses LLMs as editors to automatically refine tool-related documentation and knowledge base context. VGCO works in two stages. First, Evaluation collects real-world failure cases and identifies mismatches between tools and their context. Second, Optimization performs hierarchical editing through offline learning with structure-aware, in-context optimization. The novelty of our LLM editors has three main aspects. First, they use a hierarchical structure that naturally integrates into the tool-calling workflow. Second, they are state-aware, action-specific, and verification-guided, which constrains the search space and enables efficient, targeted improvements. Third, they enable cost-efficient sub-task specialization, either by prompt engineering large editor models or by post-training smaller editor models. Unlike prior work that emphasizes multi-turn reasoning, VGCO focuses on the single-turn, large-scale tool-calling problem and achieves significant improvements in accuracy, robustness, and generalization across LLMs.
Imbalanced Gradients in RL Post-Training of Multi-Task LLMs
Oct 22, 2025Abstract:Multi-task post-training of large language models (LLMs) is typically performed by mixing datasets from different tasks and optimizing them jointly. This approach implicitly assumes that all tasks contribute gradients of similar magnitudes; when this assumption fails, optimization becomes biased toward large-gradient tasks. In this paper, however, we show that this assumption fails in RL post-training: certain tasks produce significantly larger gradients, thus biasing updates toward those tasks. Such gradient imbalance would be justified only if larger gradients implied larger learning gains on the tasks (i.e., larger performance improvements) -- but we find this is not true. Large-gradient tasks can achieve similar or even much lower learning gains than small-gradient ones. Further analyses reveal that these gradient imbalances cannot be explained by typical training statistics such as training rewards or advantages, suggesting that they arise from the inherent differences between tasks. This cautions against naive dataset mixing and calls for future work on principled gradient-level corrections for LLMs.
Actor-Free Continuous Control via Structurally Maximizable Q-Functions
Oct 21, 2025Abstract:Value-based algorithms are a cornerstone of off-policy reinforcement learning due to their simplicity and training stability. However, their use has traditionally been restricted to discrete action spaces, as they rely on estimating Q-values for individual state-action pairs. In continuous action spaces, evaluating the Q-value over the entire action space becomes computationally infeasible. To address this, actor-critic methods are typically employed, where a critic is trained on off-policy data to estimate Q-values, and an actor is trained to maximize the critic's output. Despite their popularity, these methods often suffer from instability during training. In this work, we propose a purely value-based framework for continuous control that revisits structural maximization of Q-functions, introducing a set of key architectural and algorithmic choices to enable efficient and stable learning. We evaluate the proposed actor-free Q-learning approach on a range of standard simulation tasks, demonstrating performance and sample efficiency on par with state-of-the-art baselines, without the cost of learning a separate actor. Particularly, in environments with constrained action spaces, where the value functions are typically non-smooth, our method with structural maximization outperforms traditional actor-critic methods with gradient-based maximization. We have released our code at https://github.com/USC-Lira/Q3C.
Internalizing Self-Consistency in Language Models: Multi-Agent Consensus Alignment
Sep 18, 2025Abstract:Language Models (LMs) are inconsistent reasoners, often generating contradictory responses to identical prompts. While inference-time methods can mitigate these inconsistencies, they fail to address the core problem: LMs struggle to reliably select reasoning pathways leading to consistent outcomes under exploratory sampling. To address this, we formalize self-consistency as an intrinsic property of well-aligned reasoning models and introduce Multi-Agent Consensus Alignment (MACA), a reinforcement learning framework that post-trains models to favor reasoning trajectories aligned with their internal consensus using majority/minority outcomes from multi-agent debate. These trajectories emerge from deliberative exchanges where agents ground reasoning in peer arguments, not just aggregation of independent attempts, creating richer consensus signals than single-round majority voting. MACA enables agents to teach themselves to be more decisive and concise, and better leverage peer insights in multi-agent settings without external supervision, driving substantial improvements across self-consistency (+27.6% on GSM8K), single-agent reasoning (+23.7% on MATH), sampling-based inference (+22.4% Pass@20 on MATH), and multi-agent ensemble decision-making (+42.7% on MathQA). These findings, coupled with strong generalization to unseen benchmarks (+16.3% on GPQA, +11.6% on CommonsenseQA), demonstrate robust self-alignment that more reliably unlocks latent reasoning potential of language models.
Grounded Reinforcement Learning for Visual Reasoning
May 29, 2025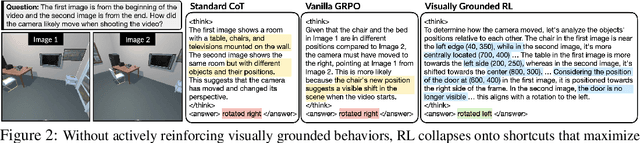
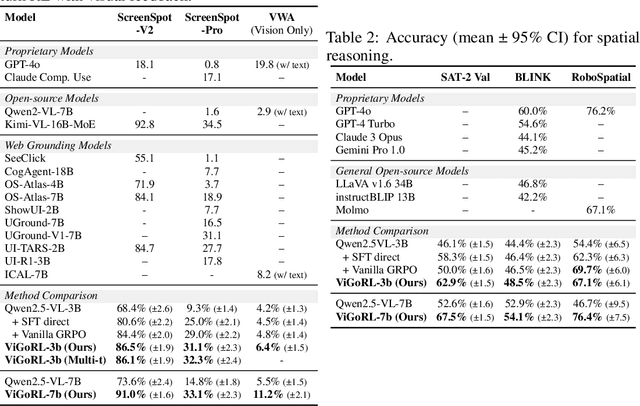
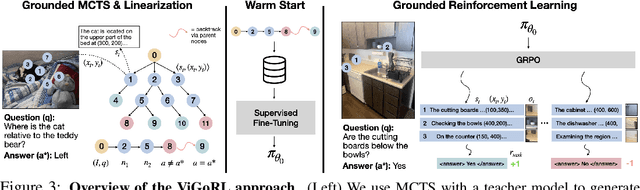
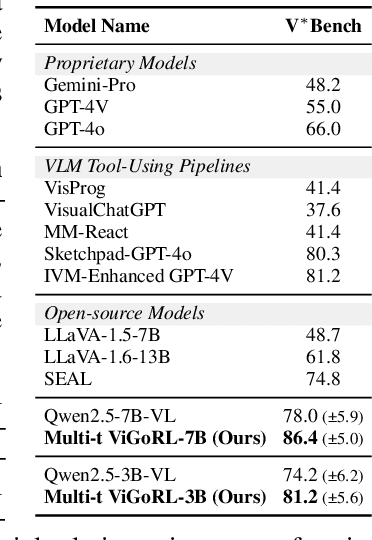
Abstract:While reinforcement learning (RL) over chains of thought has significantly advanced language models in tasks such as mathematics and coding, visual reasoning introduces added complexity by requiring models to direct visual attention, interpret perceptual inputs, and ground abstract reasoning in spatial evidence. We introduce ViGoRL (Visually Grounded Reinforcement Learning), a vision-language model trained with RL to explicitly anchor each reasoning step to specific visual coordinates. Inspired by human visual decision-making, ViGoRL learns to produce spatially grounded reasoning traces, guiding visual attention to task-relevant regions at each step. When fine-grained exploration is required, our novel multi-turn RL framework enables the model to dynamically zoom into predicted coordinates as reasoning unfolds. Across a diverse set of visual reasoning benchmarks--including SAT-2 and BLINK for spatial reasoning, V*bench for visual search, and ScreenSpot and VisualWebArena for web-based grounding--ViGoRL consistently outperforms both supervised fine-tuning and conventional RL baselines that lack explicit grounding mechanisms. Incorporating multi-turn RL with zoomed-in visual feedback significantly improves ViGoRL's performance on localizing small GUI elements and visual search, achieving 86.4% on V*Bench. Additionally, we find that grounding amplifies other visual behaviors such as region exploration, grounded subgoal setting, and visual verification. Finally, human evaluations show that the model's visual references are not only spatially accurate but also helpful for understanding model reasoning steps. Our results show that visually grounded RL is a strong paradigm for imbuing models with general-purpose visual reasoning.
polyGen: A Learning Framework for Atomic-level Polymer Structure Generation
Apr 24, 2025Abstract:Synthetic polymeric materials underpin fundamental technologies in the energy, electronics, consumer goods, and medical sectors, yet their development still suffers from prolonged design timelines. Although polymer informatics tools have supported speedup, polymer simulation protocols continue to face significant challenges: on-demand generation of realistic 3D atomic structures that respect the conformational diversity of polymer structures. Generative algorithms for 3D structures of inorganic crystals, bio-polymers, and small molecules exist, but have not addressed synthetic polymers. In this work, we introduce polyGen, the first latent diffusion model designed specifically to generate realistic polymer structures from minimal inputs such as the repeat unit chemistry alone, leveraging a molecular encoding that captures polymer connectivity throughout the architecture. Due to a scarce dataset of only 3855 DFT-optimized polymer structures, we augment our training with DFT-optimized molecular structures, showing improvement in joint learning between similar chemical structures. We also establish structure matching criteria to benchmark our approach on this novel problem. polyGen effectively generates diverse conformations of both linear chains and complex branched structures, though its performance decreases when handling repeat units with a high atom count. Given these initial results, polyGen represents a paradigm shift in atomic-level structure generation for polymer science-the first proof-of-concept for predicting realistic atomic-level polymer conformations while accounting for their intrinsic structural flexibility.
Locate 3D: Real-World Object Localization via Self-Supervised Learning in 3D
Apr 19, 2025Abstract:We present LOCATE 3D, a model for localizing objects in 3D scenes from referring expressions like "the small coffee table between the sofa and the lamp." LOCATE 3D sets a new state-of-the-art on standard referential grounding benchmarks and showcases robust generalization capabilities. Notably, LOCATE 3D operates directly on sensor observation streams (posed RGB-D frames), enabling real-world deployment on robots and AR devices. Key to our approach is 3D-JEPA, a novel self-supervised learning (SSL) algorithm applicable to sensor point clouds. It takes as input a 3D pointcloud featurized using 2D foundation models (CLIP, DINO). Subsequently, masked prediction in latent space is employed as a pretext task to aid the self-supervised learning of contextualized pointcloud features. Once trained, the 3D-JEPA encoder is finetuned alongside a language-conditioned decoder to jointly predict 3D masks and bounding boxes. Additionally, we introduce LOCATE 3D DATASET, a new dataset for 3D referential grounding, spanning multiple capture setups with over 130K annotations. This enables a systematic study of generalization capabilities as well as a stronger model.
Unifying 2D and 3D Vision-Language Understanding
Mar 13, 2025Abstract:Progress in 3D vision-language learning has been hindered by the scarcity of large-scale 3D datasets. We introduce UniVLG, a unified architecture for 2D and 3D vision-language understanding that bridges the gap between existing 2D-centric models and the rich 3D sensory data available in embodied systems. Our approach initializes most model weights from pre-trained 2D models and trains on both 2D and 3D vision-language data. We propose a novel language-conditioned mask decoder shared across 2D and 3D modalities to ground objects effectively in both RGB and RGB-D images, outperforming box-based approaches. To further reduce the domain gap between 2D and 3D, we incorporate 2D-to-3D lifting strategies, enabling UniVLG to utilize 2D data to enhance 3D performance. With these innovations, our model achieves state-of-the-art performance across multiple 3D vision-language grounding tasks, demonstrating the potential of transferring advances from 2D vision-language learning to the data-constrained 3D domain. Furthermore, co-training on both 2D and 3D data enhances performance across modalities without sacrificing 2D capabilities. By removing the reliance on 3D mesh reconstruction and ground-truth object proposals, UniVLG sets a new standard for realistic, embodied-aligned evaluation. Code and additional visualizations are available at $\href{https://univlg.github.io}{univlg.github.io}$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge