Christine Tanner
Delineating Bone Surfaces in B-Mode Images Constrained by Physics of Ultrasound Propagation
Jan 07, 2020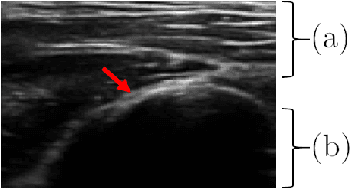
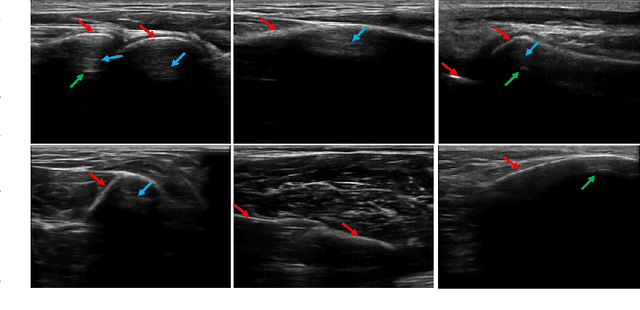
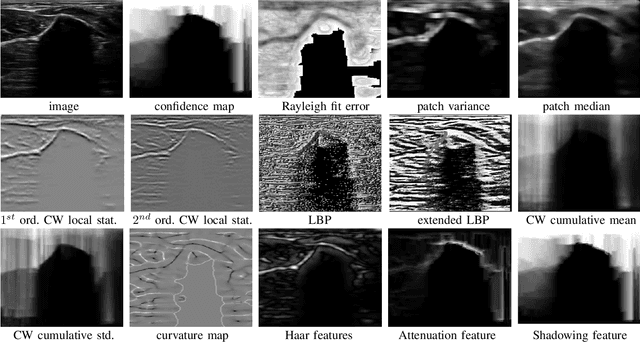
Abstract:Bone surface delineation in ultrasound is of interest due to its potential in diagnosis, surgical planning, and post-operative follow-up in orthopedics, as well as the potential of using bones as anatomical landmarks in surgical navigation. We herein propose a method to encode the physics of ultrasound propagation into a factor graph formulation for the purpose of bone surface delineation. In this graph structure, unary node potentials encode the local likelihood for being a soft tissue or acoustic-shadow (behind bone surface) region, both learned through image descriptors. Pair-wise edge potentials encode ultrasound propagation constraints of bone surfaces given their large acoustic-impedance difference. We evaluate the proposed method in comparison with four earlier approaches, on in-vivo ultrasound images collected from dorsal and volar views of the forearm. The proposed method achieves an average root-mean-square error and symmetric Hausdorff distance of 0.28mm and 1.78mm, respectively. It detects 99.9% of the annotated bone surfaces with a mean scanline error (distance to annotations) of 0.39mm.
Active Learning for Segmentation Based on Bayesian Sample Queries
Dec 22, 2019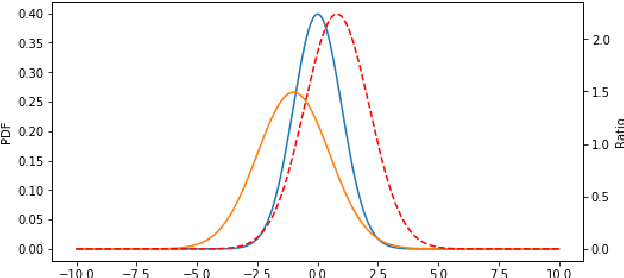

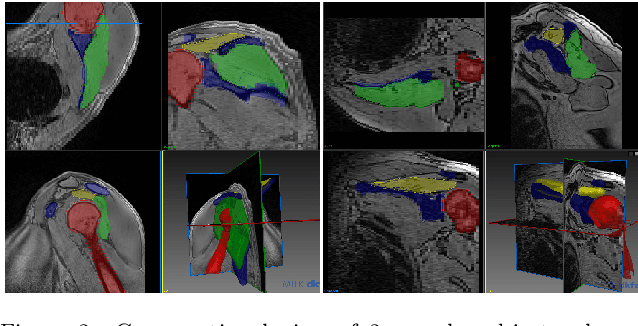
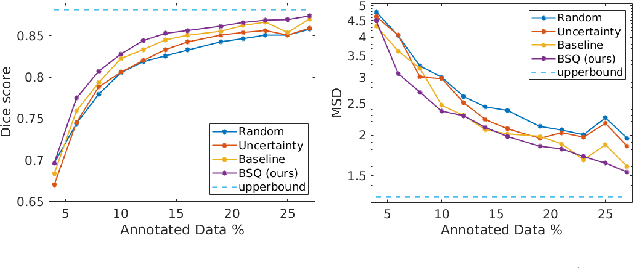
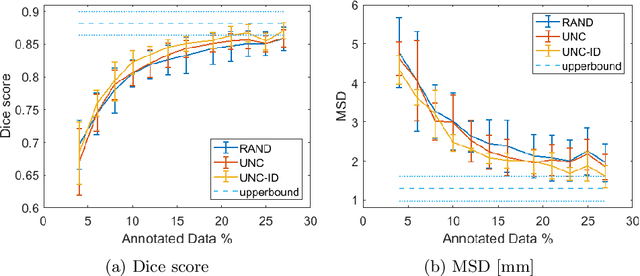
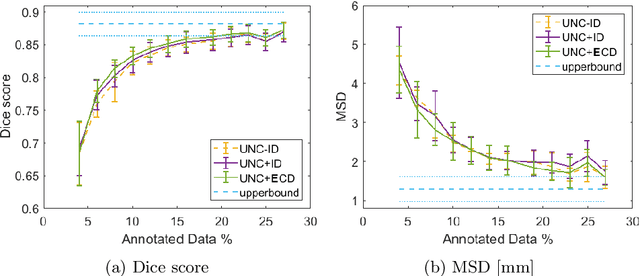
Abstract:Segmentation of anatomical structures is a fundamental image analysis task for many applications in the medical field. Deep learning methods have been shown to perform well, but for this purpose large numbers of manual annotations are needed in the first place, which necessitate prohibitive levels of resources that are often unavailable. In an active learning framework of selecting informed samples for manual labeling, expert clinician time for manual annotation can be optimally utilized, enabling the establishment of large labeled datasets for machine learning. In this paper, we propose a novel method that combines representativeness with uncertainty in order to estimate ideal samples to be annotated, iteratively from a given dataset. Our novel representativeness metric is based on Bayesian sampling, by using information-maximizing autoencoders. We conduct experiments on a shoulder magnetic resonance imaging (MRI) dataset for the segmentation of four musculoskeletal tissue classes. Quantitative results show that the annotation of representative samples selected by our proposed querying method yields an improved segmentation performance at each active learning iteration, compared to a baseline method that also employs uncertainty and representativeness metrics. For instance, with only 10% of the dataset annotated, our method reaches within 5% of Dice score expected from the upper bound scenario of all the dataset given as annotated (an impractical scenario due to resource constraints), and this gap drops down to a mere 2% when less than a fifth of the dataset samples are annotated. Such active learning approach to selecting samples to annotate enables an optimal use of the expert clinician time, being often the bottleneck in realizing machine learning solutions in medicine.
SCATGAN for Reconstruction of Ultrasound Scatterers Using Generative Adversarial Networks
Feb 01, 2019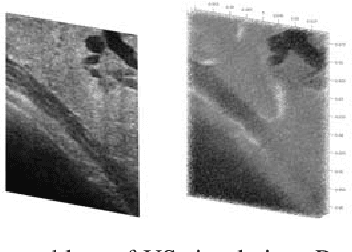
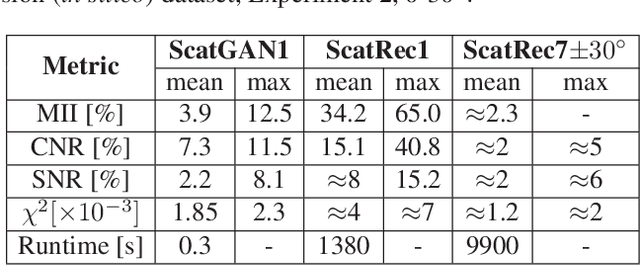
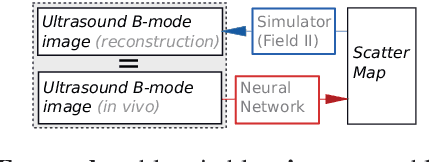
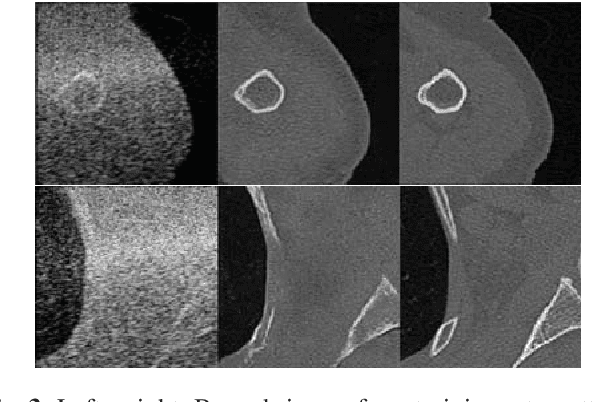
Abstract:Computational simulation of ultrasound (US) echography is essential for training sonographers. Realistic simulation of US interaction with microscopic tissue structures is often modeled by a tissue representation in the form of point scatterers, convolved with a spatially varying point spread function. This yields a realistic US B-mode speckle texture, given that a scatterer representation for a particular tissue type is readily available. This is often not the case and scatterers are nontrivial to determine. In this work we propose to estimate scatterer maps from sample US B-mode images of a tissue, by formulating this inverse mapping problem as image translation, where we learn the mapping with Generative Adversarial Networks, using a US simulation software for training. We demonstrate robust reconstruction results, invariant to US viewing and imaging settings such as imaging direction and center frequency. Our method is shown to generalize beyond the trained imaging settings, demonstrated on in-vivo US data. Our inference runs orders of magnitude faster than optimization-based techniques, enabling future extensions for reconstructing 3D B-mode volumes with only linear computational complexity.
Siamese Networks with Location Prior for Landmark Tracking in Liver Ultrasound Sequences
Jan 23, 2019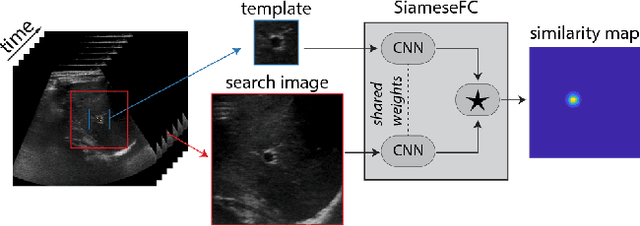
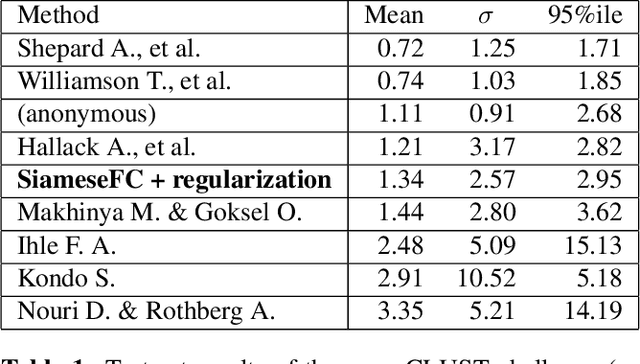
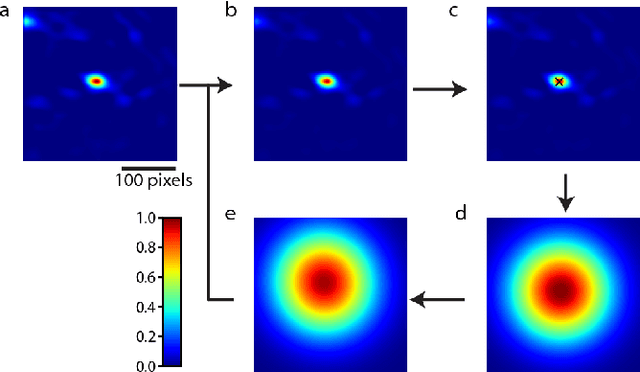
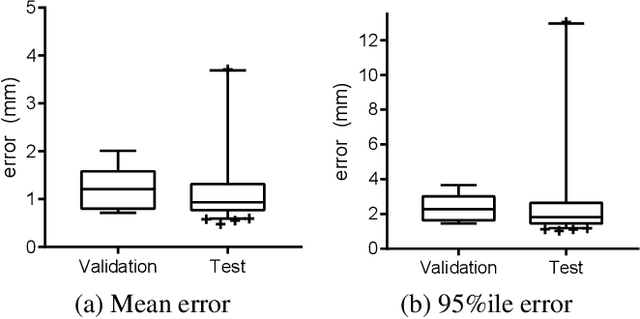
Abstract:Image-guided radiation therapy can benefit from accurate motion tracking by ultrasound imaging, in order to minimize treatment margins and radiate moving anatomical targets, e.g., due to breathing. One way to formulate this tracking problem is the automatic localization of given tracked anatomical landmarks throughout a temporal ultrasound sequence. For this, we herein propose a fully-convolutional Siamese network that learns the similarity between pairs of image regions containing the same landmark. Accordingly, it learns to localize and thus track arbitrary image features, not only predefined anatomical structures. We employ a temporal consistency model as a location prior, which we combine with the network-predicted location probability map to track a target iteratively in ultrasound sequences. We applied this method on the dataset of the Challenge on Liver Ultrasound Tracking (CLUST) with competitive results, where our work is the first to effectively apply CNNs on this tracking problem, thanks to our temporal regularization.
Iterative Interaction Training for Segmentation Editing Networks
Jul 23, 2018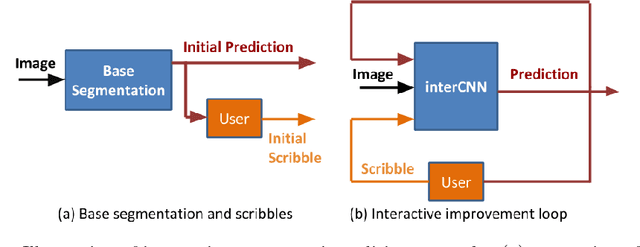
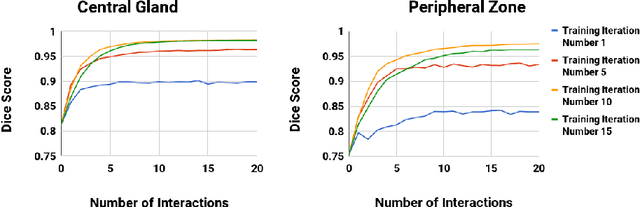
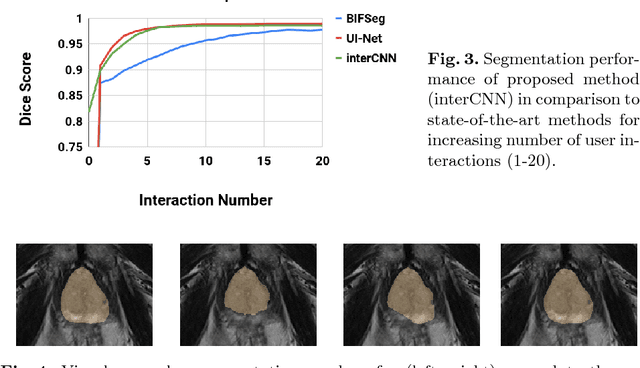
Abstract:Automatic segmentation has great potential to facilitate morphological measurements while simultaneously increasing efficiency. Nevertheless often users want to edit the segmentation to their own needs and will need different tools for this. There has been methods developed to edit segmentations of automatic methods based on the user input, primarily for binary segmentations. Here however, we present an unique training strategy for convolutional neural networks (CNNs) trained on top of an automatic method to enable interactive segmentation editing that is not limited to binary segmentation. By utilizing a robot-user during training, we closely mimic realistic use cases to achieve optimal editing performance. In addition, we show that an increase of the iterative interactions during the training process up to ten improves the segmentation editing performance substantially. Furthermore, we compare our segmentation editing CNN (interCNN) to state-of-the-art interactive segmentation algorithms and show a superior or on par performance.
Generative Adversarial Networks for MR-CT Deformable Image Registration
Jul 19, 2018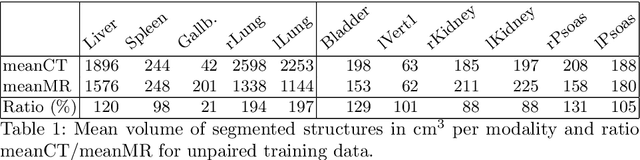
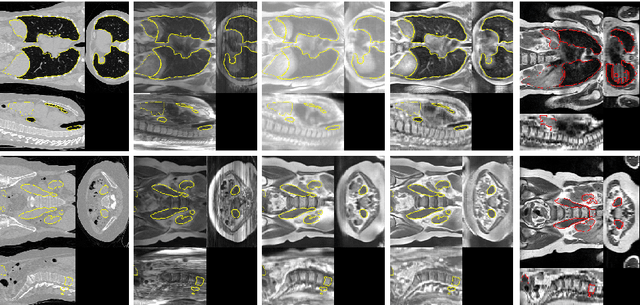
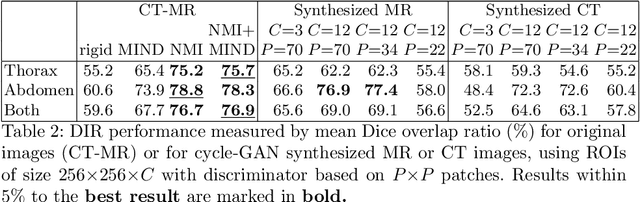
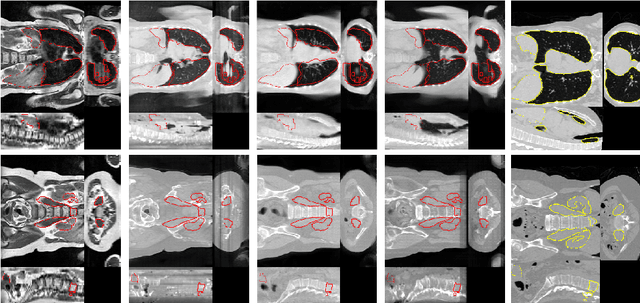
Abstract:Deformable Image Registration (DIR) of MR and CT images is one of the most challenging registration task, due to the inherent structural differences of the modalities and the missing dense ground truth. Recently cycle Generative Adversarial Networks (cycle-GANs) have been used to learn the intensity relationship between these 2 modalities for unpaired brain data. Yet its usefulness for DIR was not assessed. In this study we evaluate the DIR performance for thoracic and abdominal organs after synthesis by cycle-GAN. We show that geometric changes, which differentiate the two populations (e.g. inhale vs. exhale), are readily synthesized as well. This causes substantial problems for any application which relies on spatial correspondences being preserved between the real and the synthesized image (e.g. plan, segmentation, landmark propagation). To alleviate this problem, we investigated reducing the spatial information provided to the discriminator by decreasing the size of its receptive fields. Image synthesis was learned from 17 unpaired subjects per modality. Registration performance was evaluated with respect to manual segmentations of 11 structures for 3 subjects from the VISERAL challenge. State-of-the-art DIR methods based on Normalized Mutual Information (NMI), Modality Independent Neighborhood Descriptor (MIND) and their novel combination achieved a mean segmentation overlap ratio of 76.7, 67.7, 76.9%, respectively. This dropped to 69.1% or less when registering images synthesized by cycle-GAN based on local correlation, due to the poor performance on the thoracic region, where large lung volume changes were synthesized. Performance for the abdominal region was similar to that of CT-MRI NMI registration (77.4 vs. 78.8%) when using 3D synthesizing MRIs (12 slices) and medium sized receptive fields for the discriminator.
Active Learning for Segmentation by Optimizing Content Information for Maximal Entropy
Jul 18, 2018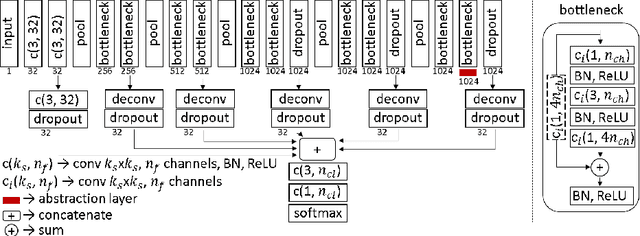

Abstract:Segmentation is essential for medical image analysis tasks such as intervention planning, therapy guidance, diagnosis, treatment decisions. Deep learning is becoming increasingly prominent for segmentation, where the lack of annotations, however, often becomes the main limitation. Due to privacy concerns and ethical considerations, most medical datasets are created, curated, and allow access only locally. Furthermore, current deep learning methods are often suboptimal in translating anatomical knowledge between different medical imaging modalities. Active learning can be used to select an informed set of image samples to request for manual annotation, in order to best utilize the limited annotation time of clinical experts for optimal outcomes, which we focus on in this work. Our contributions herein are two fold: (1) we enforce domain-representativeness of selected samples using a proposed penalization scheme to maximize information at the network abstraction layer, and (2) we propose a Borda-count based sample querying scheme for selecting samples for segmentation. Comparative experiments with baseline approaches show that the samples queried with our proposed method, where both above contributions are combined, result in significantly improved segmentation performance for this active learning task.
Temporal Interpolation via Motion Field Prediction
Apr 12, 2018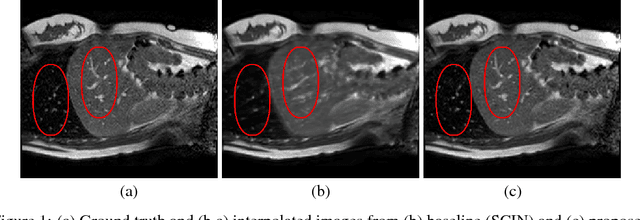
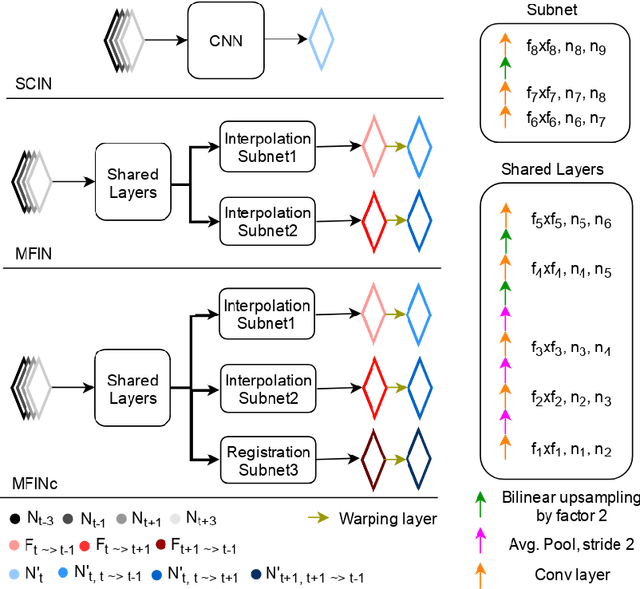
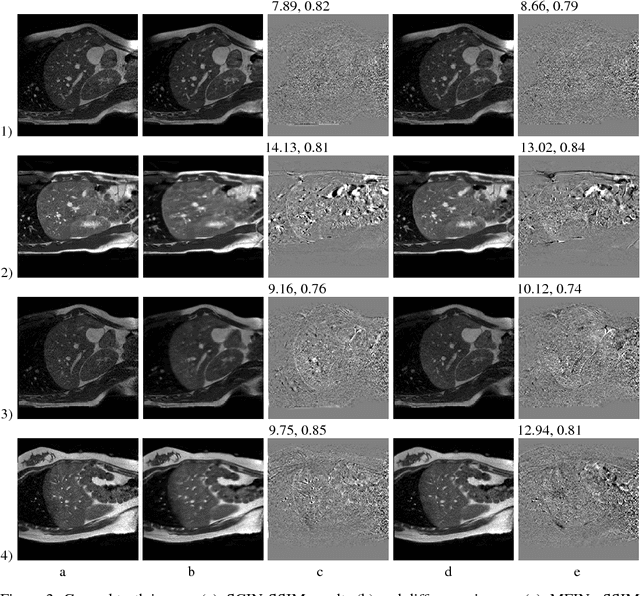
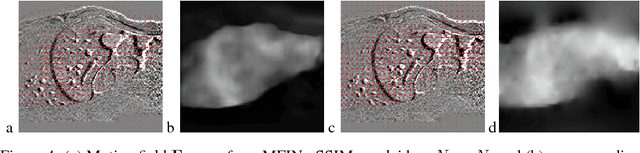
Abstract:Navigated 2D multi-slice dynamic Magnetic Resonance (MR) imaging enables high contrast 4D MR imaging during free breathing and provides in-vivo observations for treatment planning and guidance. Navigator slices are vital for retrospective stacking of 2D data slices in this method. However, they also prolong the acquisition sessions. Temporal interpolation of navigator slices an be used to reduce the number of navigator acquisitions without degrading specificity in stacking. In this work, we propose a convolutional neural network (CNN) based method for temporal interpolation via motion field prediction. The proposed formulation incorporates the prior knowledge that a motion field underlies changes in the image intensities over time. Previous approaches that interpolate directly in the intensity space are prone to produce blurry images or even remove structures in the images. Our method avoids such problems and faithfully preserves the information in the image. Further, an important advantage of our formulation is that it provides an unsupervised estimation of bi-directional motion fields. We show that these motion fields can be used to halve the number of registrations required during 4D reconstruction, thus substantially reducing the reconstruction time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge