Andrawes Al Bahou
SCATGAN for Reconstruction of Ultrasound Scatterers Using Generative Adversarial Networks
Feb 01, 2019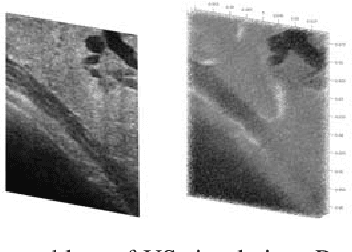
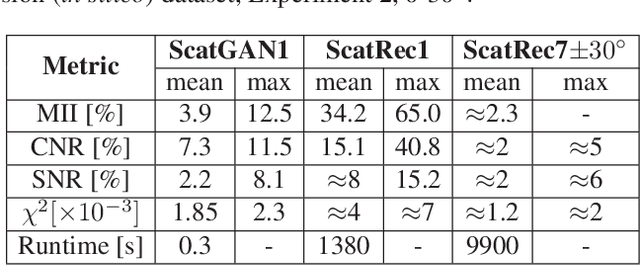
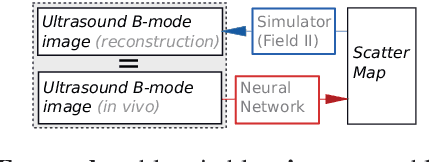
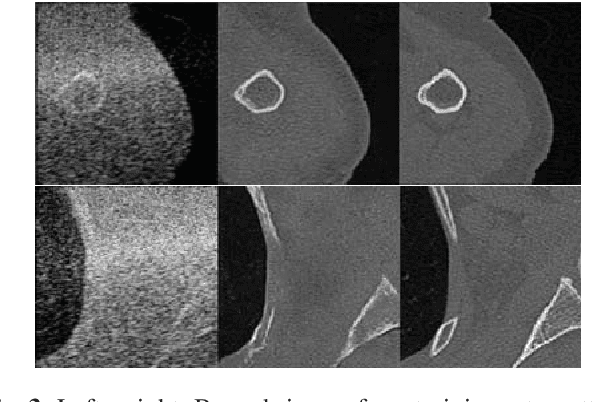
Abstract:Computational simulation of ultrasound (US) echography is essential for training sonographers. Realistic simulation of US interaction with microscopic tissue structures is often modeled by a tissue representation in the form of point scatterers, convolved with a spatially varying point spread function. This yields a realistic US B-mode speckle texture, given that a scatterer representation for a particular tissue type is readily available. This is often not the case and scatterers are nontrivial to determine. In this work we propose to estimate scatterer maps from sample US B-mode images of a tissue, by formulating this inverse mapping problem as image translation, where we learn the mapping with Generative Adversarial Networks, using a US simulation software for training. We demonstrate robust reconstruction results, invariant to US viewing and imaging settings such as imaging direction and center frequency. Our method is shown to generalize beyond the trained imaging settings, demonstrated on in-vivo US data. Our inference runs orders of magnitude faster than optimization-based techniques, enabling future extensions for reconstructing 3D B-mode volumes with only linear computational complexity.
XNORBIN: A 95 TOp/s/W Hardware Accelerator for Binary Convolutional Neural Networks
Mar 05, 2018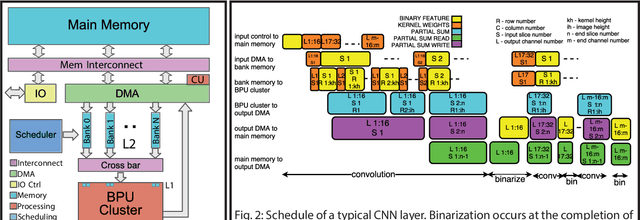
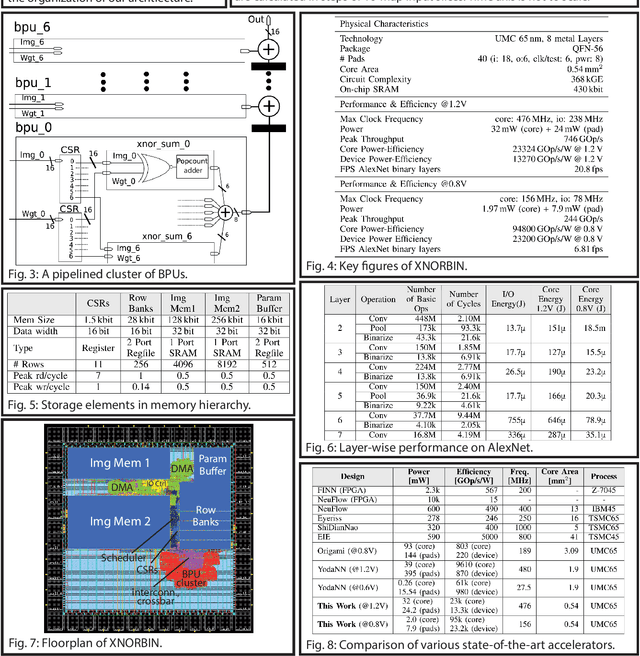
Abstract:Deploying state-of-the-art CNNs requires power-hungry processors and off-chip memory. This precludes the implementation of CNNs in low-power embedded systems. Recent research shows CNNs sustain extreme quantization, binarizing their weights and intermediate feature maps, thereby saving 8-32\x memory and collapsing energy-intensive sum-of-products into XNOR-and-popcount operations. We present XNORBIN, an accelerator for binary CNNs with computation tightly coupled to memory for aggressive data reuse. Implemented in UMC 65nm technology XNORBIN achieves an energy efficiency of 95 TOp/s/W and an area efficiency of 2.0 TOp/s/MGE at 0.8 V.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge