Romy Profanter
Generative Adversarial Networks for MR-CT Deformable Image Registration
Jul 19, 2018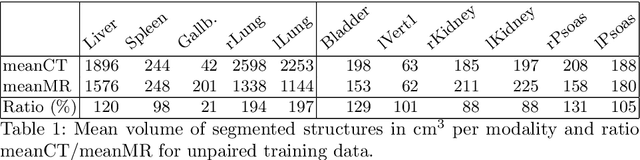
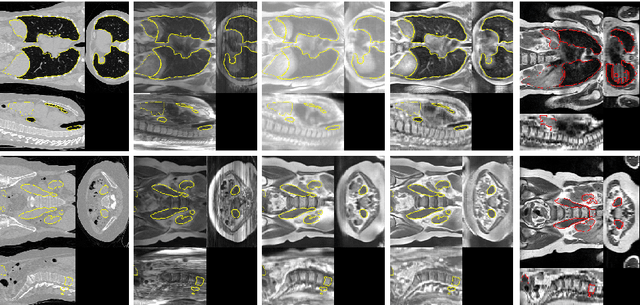
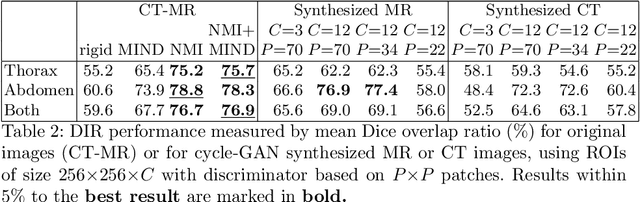
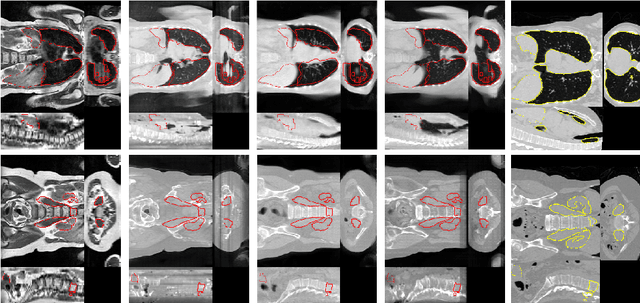
Abstract:Deformable Image Registration (DIR) of MR and CT images is one of the most challenging registration task, due to the inherent structural differences of the modalities and the missing dense ground truth. Recently cycle Generative Adversarial Networks (cycle-GANs) have been used to learn the intensity relationship between these 2 modalities for unpaired brain data. Yet its usefulness for DIR was not assessed. In this study we evaluate the DIR performance for thoracic and abdominal organs after synthesis by cycle-GAN. We show that geometric changes, which differentiate the two populations (e.g. inhale vs. exhale), are readily synthesized as well. This causes substantial problems for any application which relies on spatial correspondences being preserved between the real and the synthesized image (e.g. plan, segmentation, landmark propagation). To alleviate this problem, we investigated reducing the spatial information provided to the discriminator by decreasing the size of its receptive fields. Image synthesis was learned from 17 unpaired subjects per modality. Registration performance was evaluated with respect to manual segmentations of 11 structures for 3 subjects from the VISERAL challenge. State-of-the-art DIR methods based on Normalized Mutual Information (NMI), Modality Independent Neighborhood Descriptor (MIND) and their novel combination achieved a mean segmentation overlap ratio of 76.7, 67.7, 76.9%, respectively. This dropped to 69.1% or less when registering images synthesized by cycle-GAN based on local correlation, due to the poor performance on the thoracic region, where large lung volume changes were synthesized. Performance for the abdominal region was similar to that of CT-MRI NMI registration (77.4 vs. 78.8%) when using 3D synthesizing MRIs (12 slices) and medium sized receptive fields for the discriminator.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge