Ender Konukoglu
for the Alzheimer's Disease Neuroimaging Initiative
A Single-Parameter Factor-Graph Image Prior
Jan 13, 2026Abstract:We propose a novel piecewise smooth image model with piecewise constant local parameters that are automatically adapted to each image. Technically, the model is formulated in terms of factor graphs with NUP (normal with unknown parameters) priors, and the pertinent computations amount to iterations of conjugate-gradient steps and Gaussian message passing. The proposed model and algorithms are demonstrated with applications to denoising and contrast enhancement.
A Multi-Centric Anthropomorphic 3D CT Phantom-Based Benchmark Dataset for Harmonization
Jul 02, 2025Abstract:Artificial intelligence (AI) has introduced numerous opportunities for human assistance and task automation in medicine. However, it suffers from poor generalization in the presence of shifts in the data distribution. In the context of AI-based computed tomography (CT) analysis, significant data distribution shifts can be caused by changes in scanner manufacturer, reconstruction technique or dose. AI harmonization techniques can address this problem by reducing distribution shifts caused by various acquisition settings. This paper presents an open-source benchmark dataset containing CT scans of an anthropomorphic phantom acquired with various scanners and settings, which purpose is to foster the development of AI harmonization techniques. Using a phantom allows fixing variations attributed to inter- and intra-patient variations. The dataset includes 1378 image series acquired with 13 scanners from 4 manufacturers across 8 institutions using a harmonized protocol as well as several acquisition doses. Additionally, we present a methodology, baseline results and open-source code to assess image- and feature-level stability and liver tissue classification, promoting the development of AI harmonization strategies.
Exploiting Temporal State Space Sharing for Video Semantic Segmentation
Mar 26, 2025Abstract:Video semantic segmentation (VSS) plays a vital role in understanding the temporal evolution of scenes. Traditional methods often segment videos frame-by-frame or in a short temporal window, leading to limited temporal context, redundant computations, and heavy memory requirements. To this end, we introduce a Temporal Video State Space Sharing (TV3S) architecture to leverage Mamba state space models for temporal feature sharing. Our model features a selective gating mechanism that efficiently propagates relevant information across video frames, eliminating the need for a memory-heavy feature pool. By processing spatial patches independently and incorporating shifted operation, TV3S supports highly parallel computation in both training and inference stages, which reduces the delay in sequential state space processing and improves the scalability for long video sequences. Moreover, TV3S incorporates information from prior frames during inference, achieving long-range temporal coherence and superior adaptability to extended sequences. Evaluations on the VSPW and Cityscapes datasets reveal that our approach outperforms current state-of-the-art methods, establishing a new standard for VSS with consistent results across long video sequences. By achieving a good balance between accuracy and efficiency, TV3S shows a significant advancement in spatiotemporal modeling, paving the way for efficient video analysis. The code is publicly available at https://github.com/Ashesham/TV3S.git.
CamSAM2: Segment Anything Accurately in Camouflaged Videos
Mar 26, 2025Abstract:Video camouflaged object segmentation (VCOS), aiming at segmenting camouflaged objects that seamlessly blend into their environment, is a fundamental vision task with various real-world applications. With the release of SAM2, video segmentation has witnessed significant progress. However, SAM2's capability of segmenting camouflaged videos is suboptimal, especially when given simple prompts such as point and box. To address the problem, we propose Camouflaged SAM2 (CamSAM2), which enhances SAM2's ability to handle camouflaged scenes without modifying SAM2's parameters. Specifically, we introduce a decamouflaged token to provide the flexibility of feature adjustment for VCOS. To make full use of fine-grained and high-resolution features from the current frame and previous frames, we propose implicit object-aware fusion (IOF) and explicit object-aware fusion (EOF) modules, respectively. Object prototype generation (OPG) is introduced to abstract and memorize object prototypes with informative details using high-quality features from previous frames. Extensive experiments are conducted to validate the effectiveness of our approach. While CamSAM2 only adds negligible learnable parameters to SAM2, it substantially outperforms SAM2 on three VCOS datasets, especially achieving 12.2 mDice gains with click prompt on MoCA-Mask and 19.6 mDice gains with mask prompt on SUN-SEG-Hard, with Hiera-T as the backbone. The code will be available at https://github.com/zhoustan/CamSAM2.
Learning to segment anatomy and lesions from disparately labeled sources in brain MRI
Mar 25, 2025Abstract:Segmenting healthy tissue structures alongside lesions in brain Magnetic Resonance Images (MRI) remains a challenge for today's algorithms due to lesion-caused disruption of the anatomy and lack of jointly labeled training datasets, where both healthy tissues and lesions are labeled on the same images. In this paper, we propose a method that is robust to lesion-caused disruptions and can be trained from disparately labeled training sets, i.e., without requiring jointly labeled samples, to automatically segment both. In contrast to prior work, we decouple healthy tissue and lesion segmentation in two paths to leverage multi-sequence acquisitions and merge information with an attention mechanism. During inference, an image-specific adaptation reduces adverse influences of lesion regions on healthy tissue predictions. During training, the adaptation is taken into account through meta-learning and co-training is used to learn from disparately labeled training images. Our model shows an improved performance on several anatomical structures and lesions on a publicly available brain glioblastoma dataset compared to the state-of-the-art segmentation methods.
Generalized Few-shot 3D Point Cloud Segmentation with Vision-Language Model
Mar 20, 2025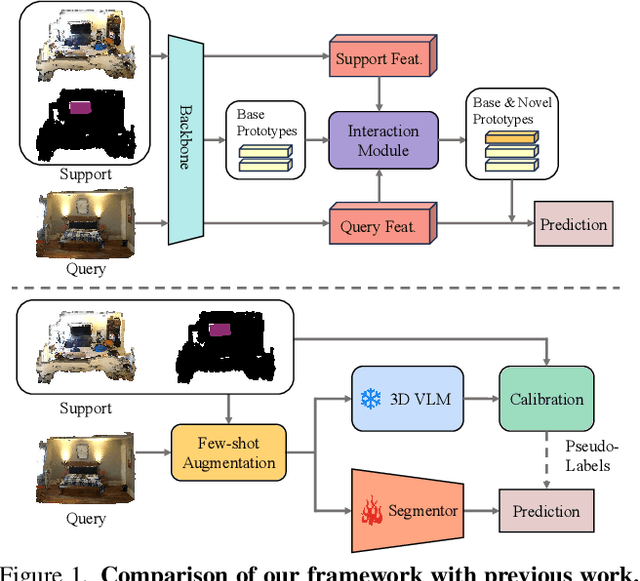
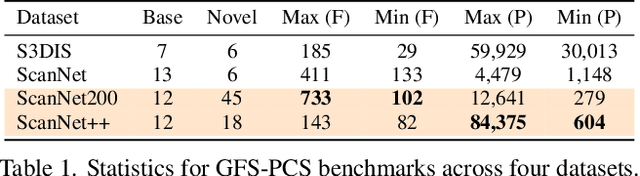
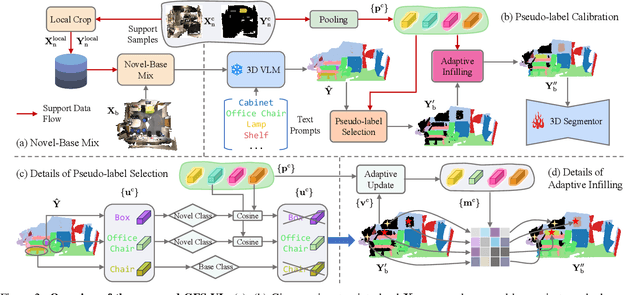
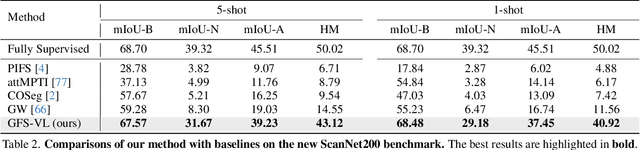
Abstract:Generalized few-shot 3D point cloud segmentation (GFS-PCS) adapts models to new classes with few support samples while retaining base class segmentation. Existing GFS-PCS methods enhance prototypes via interacting with support or query features but remain limited by sparse knowledge from few-shot samples. Meanwhile, 3D vision-language models (3D VLMs), generalizing across open-world novel classes, contain rich but noisy novel class knowledge. In this work, we introduce a GFS-PCS framework that synergizes dense but noisy pseudo-labels from 3D VLMs with precise yet sparse few-shot samples to maximize the strengths of both, named GFS-VL. Specifically, we present a prototype-guided pseudo-label selection to filter low-quality regions, followed by an adaptive infilling strategy that combines knowledge from pseudo-label contexts and few-shot samples to adaptively label the filtered, unlabeled areas. Additionally, we design a novel-base mix strategy to embed few-shot samples into training scenes, preserving essential context for improved novel class learning. Moreover, recognizing the limited diversity in current GFS-PCS benchmarks, we introduce two challenging benchmarks with diverse novel classes for comprehensive generalization evaluation. Experiments validate the effectiveness of our framework across models and datasets. Our approach and benchmarks provide a solid foundation for advancing GFS-PCS in the real world. The code is at https://github.com/ZhaochongAn/GFS-VL
Conformal forecasting for surgical instrument trajectory
Mar 06, 2025


Abstract:Forecasting surgical instrument trajectories and predicting the next surgical action recently started to attract attention from the research community. Both these tasks are crucial for automation and assistance in endoscopy surgery. Given the safety-critical nature of these tasks, reliable uncertainty quantification is essential. Conformal prediction is a fast-growing and widely recognized framework for uncertainty estimation in machine learning and computer vision, offering distribution-free, theoretically valid prediction intervals. In this work, we explore the application of standard conformal prediction and conformalized quantile regression to estimate uncertainty in forecasting surgical instrument motion, i.e., predicting direction and magnitude of surgical instruments' future motion. We analyze and compare their coverage and interval sizes, assessing the impact of multiple hypothesis testing and correction methods. Additionally, we show how these techniques can be employed to produce useful uncertainty heatmaps. To the best of our knowledge, this is the first study applying conformal prediction to surgical guidance, marking an initial step toward constructing principled prediction intervals with formal coverage guarantees in this domain.
Tera-MIND: Tera-scale mouse brain simulation via spatial mRNA-guided diffusion
Mar 04, 2025Abstract:Holistic 3D modeling of molecularly defined brain structures is crucial for understanding complex brain functions. Emerging tissue profiling technologies enable the construction of a comprehensive atlas of the mammalian brain with sub-cellular resolution and spatially resolved gene expression data. However, such tera-scale volumetric datasets present significant computational challenges in understanding complex brain functions within their native 3D spatial context. Here, we propose the novel generative approach $\textbf{Tera-MIND}$, which can simulate $\textbf{Tera}$-scale $\textbf{M}$ouse bra$\textbf{IN}$s in 3D using a patch-based and boundary-aware $\textbf{D}$iffusion model. Taking spatial transcriptomic data as the conditional input, we generate virtual mouse brains with comprehensive cellular morphological detail at teravoxel scale. Through the lens of 3D $gene$-$gene$ self-attention, we identify spatial molecular interactions for key transcriptomic pathways in the murine brain, exemplified by glutamatergic and dopaminergic neuronal systems. Importantly, these $in$-$silico$ biological findings are consistent and reproducible across three tera-scale virtual mouse brains. Therefore, Tera-MIND showcases a promising path toward efficient and generative simulations of whole organ systems for biomedical research. Project website: https://musikisomorphie.github.io/Tera-MIND.html
Anatomy Might Be All You Need: Forecasting What to Do During Surgery
Jan 31, 2025Abstract:Surgical guidance can be delivered in various ways. In neurosurgery, spatial guidance and orientation are predominantly achieved through neuronavigation systems that reference pre-operative MRI scans. Recently, there has been growing interest in providing live guidance by analyzing video feeds from tools such as endoscopes. Existing approaches, including anatomy detection, orientation feedback, phase recognition, and visual question-answering, primarily focus on aiding surgeons in assessing the current surgical scene. This work aims to provide guidance on a finer scale, aiming to provide guidance by forecasting the trajectory of the surgical instrument, essentially addressing the question of what to do next. To address this task, we propose a model that not only leverages the historical locations of surgical instruments but also integrates anatomical features. Importantly, our work does not rely on explicit ground truth labels for instrument trajectories. Instead, the ground truth is generated by a detection model trained to detect both anatomical structures and instruments within surgical videos of a comprehensive dataset containing pituitary surgery videos. By analyzing the interaction between anatomy and instrument movements in these videos and forecasting future instrument movements, we show that anatomical features are a valuable asset in addressing this challenging task. To the best of our knowledge, this work is the first attempt to address this task for manually operated surgeries.
CityLoc: 6 DoF Localization of Text Descriptions in Large-Scale Scenes with Gaussian Representation
Jan 15, 2025Abstract:Localizing text descriptions in large-scale 3D scenes is inherently an ambiguous task. This nonetheless arises while describing general concepts, e.g. all traffic lights in a city. To facilitate reasoning based on such concepts, text localization in the form of distribution is required. In this paper, we generate the distribution of the camera poses conditioned upon the textual description. To facilitate such generation, we propose a diffusion-based architecture that conditionally diffuses the noisy 6DoF camera poses to their plausible locations. The conditional signals are derived from the text descriptions, using the pre-trained text encoders. The connection between text descriptions and pose distribution is established through pretrained Vision-Language-Model, i.e. CLIP. Furthermore, we demonstrate that the candidate poses for the distribution can be further refined by rendering potential poses using 3D Gaussian splatting, guiding incorrectly posed samples towards locations that better align with the textual description, through visual reasoning. We demonstrate the effectiveness of our method by comparing it with both standard retrieval methods and learning-based approaches. Our proposed method consistently outperforms these baselines across all five large-scale datasets. Our source code and dataset will be made publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge