Markus Reischl
Learning to Detect Label Errors by Making Them: A Method for Segmentation and Object Detection Datasets
Aug 25, 2025Abstract:Recently, detection of label errors and improvement of label quality in datasets for supervised learning tasks has become an increasingly important goal in both research and industry. The consequences of incorrectly annotated data include reduced model performance, biased benchmark results, and lower overall accuracy. Current state-of-the-art label error detection methods often focus on a single computer vision task and, consequently, a specific type of dataset, containing, for example, either bounding boxes or pixel-wise annotations. Furthermore, previous methods are not learning-based. In this work, we overcome this research gap. We present a unified method for detecting label errors in object detection, semantic segmentation, and instance segmentation datasets. In a nutshell, our approach - learning to detect label errors by making them - works as follows: we inject different kinds of label errors into the ground truth. Then, the detection of label errors, across all mentioned primary tasks, is framed as an instance segmentation problem based on a composite input. In our experiments, we compare the label error detection performance of our method with various baselines and state-of-the-art approaches of each task's domain on simulated label errors across multiple tasks, datasets, and base models. This is complemented by a generalization study on real-world label errors. Additionally, we release 459 real label errors identified in the Cityscapes dataset and provide a benchmark for real label error detection in Cityscapes.
Large Means Left: Political Bias in Large Language Models Increases with Their Number of Parameters
May 07, 2025Abstract:With the increasing prevalence of artificial intelligence, careful evaluation of inherent biases needs to be conducted to form the basis for alleviating the effects these predispositions can have on users. Large language models (LLMs) are predominantly used by many as a primary source of information for various topics. LLMs frequently make factual errors, fabricate data (hallucinations), or present biases, exposing users to misinformation and influencing opinions. Educating users on their risks is key to responsible use, as bias, unlike hallucinations, cannot be caught through data verification. We quantify the political bias of popular LLMs in the context of the recent vote of the German Bundestag using the score produced by the Wahl-O-Mat. This metric measures the alignment between an individual's political views and the positions of German political parties. We compare the models' alignment scores to identify factors influencing their political preferences. Doing so, we discover a bias toward left-leaning parties, most dominant in larger LLMs. Also, we find that the language we use to communicate with the models affects their political views. Additionally, we analyze the influence of a model's origin and release date and compare the results to the outcome of the recent vote of the Bundestag. Our results imply that LLMs are prone to exhibiting political bias. Large corporations with the necessary means to develop LLMs, thus, knowingly or unknowingly, have a responsibility to contain these biases, as they can influence each voter's decision-making process and inform public opinion in general and at scale.
Energy-Based Prior Latent Space Diffusion model for Reconstruction of Lumbar Vertebrae from Thick Slice MRI
Nov 30, 2024Abstract:Lumbar spine problems are ubiquitous, motivating research into targeted imaging for treatment planning and guided interventions. While high resolution and high contrast CT has been the modality of choice, MRI can capture both bone and soft tissue without the ionizing radiation of CT albeit longer acquisition time. The critical trade-off between contrast quality and acquisition time has motivated 'thick slice MRI', which prioritises faster imaging with high in-plane resolution but variable contrast and low through-plane resolution. We investigate a recently developed post-acquisition pipeline which segments vertebrae from thick-slice acquisitions and uses a variational autoencoder to enhance quality after an initial 3D reconstruction. We instead propose a latent space diffusion energy-based prior to leverage diffusion models, which exhibit high-quality image generation. Crucially, we mitigate their high computational cost and low sample efficiency by learning an energy-based latent representation to perform the diffusion processes. Our resulting method outperforms existing approaches across metrics including Dice and VS scores, and more faithfully captures 3D features.
Improving 3D deep learning segmentation with biophysically motivated cell synthesis
Aug 29, 2024Abstract:Biomedical research increasingly relies on 3D cell culture models and AI-based analysis can potentially facilitate a detailed and accurate feature extraction on a single-cell level. However, this requires for a precise segmentation of 3D cell datasets, which in turn demands high-quality ground truth for training. Manual annotation, the gold standard for ground truth data, is too time-consuming and thus not feasible for the generation of large 3D training datasets. To address this, we present a novel framework for generating 3D training data, which integrates biophysical modeling for realistic cell shape and alignment. Our approach allows the in silico generation of coherent membrane and nuclei signals, that enable the training of segmentation models utilizing both channels for improved performance. Furthermore, we present a new GAN training scheme that generates not only image data but also matching labels. Quantitative evaluation shows superior performance of biophysical motivated synthetic training data, even outperforming manual annotation and pretrained models. This underscores the potential of incorporating biophysical modeling for enhancing synthetic training data quality.
Assessing Political Bias in Large Language Models
May 17, 2024



Abstract:The assessment of societal biases within Large Language Models (LLMs) has emerged as a critical concern in the contemporary discourse surrounding Artificial Intelligence (AI) ethics and their impact. Especially, recognizing and considering political biases is important for practical applications to gain a deeper understanding of the possibilities and behaviors and to prevent unwanted statements. As the upcoming elections of the European Parliament will not remain unaffected by LLMs, we evaluate the bias of the current most popular open-source models concerning political issues within the European Union (EU) from a German perspective. To do so, we use the "Wahl-O-Mat", a voting advice application used in Germany, to determine which political party is the most aligned for the respective LLM. We show that larger models, such as Llama3-70B, tend to align more closely with left-leaning political parties like GR\"UNE and Volt, while smaller models often remain neutral, particularly in English. This highlights the nuanced behavior of LLMs and the importance of language in shaping their political stances. Our findings underscore the importance of rigorously assessing and addressing societal bias in LLMs to safeguard the integrity and fairness of applications that employ the power of modern machine learning methods.
MLOps for Scarce Image Data: A Use Case in Microscopic Image Analysis
Oct 04, 2023Abstract:Nowadays, Machine Learning (ML) is experiencing tremendous popularity that has never been seen before. The operationalization of ML models is governed by a set of concepts and methods referred to as Machine Learning Operations (MLOps). Nevertheless, researchers, as well as professionals, often focus more on the automation aspect and neglect the continuous deployment and monitoring aspects of MLOps. As a result, there is a lack of continuous learning through the flow of feedback from production to development, causing unexpected model deterioration over time due to concept drifts, particularly when dealing with scarce data. This work explores the complete application of MLOps in the context of scarce data analysis. The paper proposes a new holistic approach to enhance biomedical image analysis. Our method includes: a fingerprinting process that enables selecting the best models, datasets, and model development strategy relative to the image analysis task at hand; an automated model development stage; and a continuous deployment and monitoring process to ensure continuous learning. For preliminary results, we perform a proof of concept for fingerprinting in microscopic image datasets.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
EasyMLServe: Easy Deployment of REST Machine Learning Services
Nov 26, 2022



Abstract:Various research domains use machine learning approaches because they can solve complex tasks by learning from data. Deploying machine learning models, however, is not trivial and developers have to implement complete solutions which are often installed locally and include Graphical User Interfaces (GUIs). Distributing software to various users on-site has several problems. Therefore, we propose a concept to deploy software in the cloud. There are several frameworks available based on Representational State Transfer (REST) which can be used to implement cloud-based machine learning services. However, machine learning services for scientific users have special requirements that state-of-the-art REST frameworks do not cover completely. We contribute an EasyMLServe software framework to deploy machine learning services in the cloud using REST interfaces and generic local or web-based GUIs. Furthermore, we apply our framework on two real-world applications, \ie, energy time-series forecasting and cell instance segmentation. The EasyMLServe framework and the use cases are available on GitHub.
A universal synthetic dataset for machine learning on spectroscopic data
Jun 14, 2022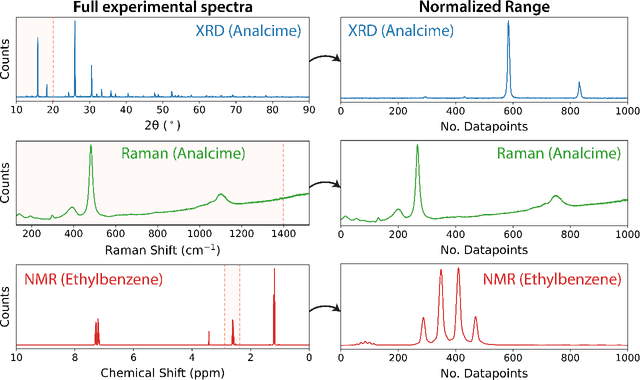
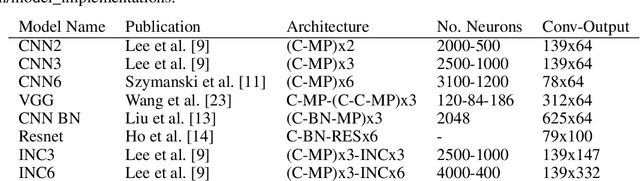
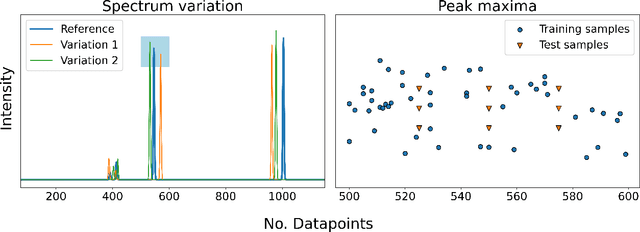
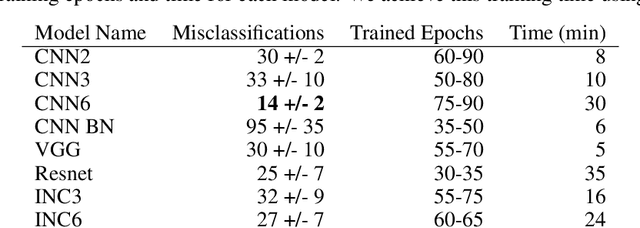
Abstract:To assist in the development of machine learning methods for automated classification of spectroscopic data, we have generated a universal synthetic dataset that can be used for model validation. This dataset contains artificial spectra designed to represent experimental measurements from techniques including X-ray diffraction, nuclear magnetic resonance, and Raman spectroscopy. The dataset generation process features customizable parameters, such as scan length and peak count, which can be adjusted to fit the problem at hand. As an initial benchmark, we simulated a dataset containing 35,000 spectra based on 500 unique classes. To automate the classification of this data, eight different machine learning architectures were evaluated. From the results, we shed light on which factors are most critical to achieve optimal performance for the classification task. The scripts used to generate synthetic spectra, as well as our benchmark dataset and evaluation routines, are made publicly available to aid in the development of improved machine learning models for spectroscopic analysis.
ciscNet -- A Single-Branch Cell Instance Segmentation and Classification Network
Feb 25, 2022


Abstract:Automated cell nucleus segmentation and classification are required to assist pathologists in their decision making. The Colon Nuclei Identification and Counting Challenge 2022 (CoNIC Challenge 2022) supports the development and comparability of segmentation and classification methods for histopathological images. In this contribution, we describe our CoNIC Challenge 2022 method ciscNet to segment, classify and count cell nuclei, and report preliminary evaluation results. Our code is available at https://git.scc.kit.edu/ciscnet/ciscnet-conic-2022.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge