Viktor H. Koelzer
Revisiting Automatic Data Curation for Vision Foundation Models in Digital Pathology
Mar 24, 2025Abstract:Vision foundation models (FMs) are accelerating the development of digital pathology algorithms and transforming biomedical research. These models learn, in a self-supervised manner, to represent histological features in highly heterogeneous tiles extracted from whole-slide images (WSIs) of real-world patient samples. The performance of these FMs is significantly influenced by the size, diversity, and balance of the pre-training data. However, data selection has been primarily guided by expert knowledge at the WSI level, focusing on factors such as disease classification and tissue types, while largely overlooking the granular details available at the tile level. In this paper, we investigate the potential of unsupervised automatic data curation at the tile-level, taking into account 350 million tiles. Specifically, we apply hierarchical clustering trees to pre-extracted tile embeddings, allowing us to sample balanced datasets uniformly across the embedding space of the pretrained FM. We further identify these datasets are subject to a trade-off between size and balance, potentially compromising the quality of representations learned by FMs, and propose tailored batch sampling strategies to mitigate this effect. We demonstrate the effectiveness of our method through improved performance on a diverse range of clinically relevant downstream tasks.
Tera-MIND: Tera-scale mouse brain simulation via spatial mRNA-guided diffusion
Mar 04, 2025Abstract:Holistic 3D modeling of molecularly defined brain structures is crucial for understanding complex brain functions. Emerging tissue profiling technologies enable the construction of a comprehensive atlas of the mammalian brain with sub-cellular resolution and spatially resolved gene expression data. However, such tera-scale volumetric datasets present significant computational challenges in understanding complex brain functions within their native 3D spatial context. Here, we propose the novel generative approach $\textbf{Tera-MIND}$, which can simulate $\textbf{Tera}$-scale $\textbf{M}$ouse bra$\textbf{IN}$s in 3D using a patch-based and boundary-aware $\textbf{D}$iffusion model. Taking spatial transcriptomic data as the conditional input, we generate virtual mouse brains with comprehensive cellular morphological detail at teravoxel scale. Through the lens of 3D $gene$-$gene$ self-attention, we identify spatial molecular interactions for key transcriptomic pathways in the murine brain, exemplified by glutamatergic and dopaminergic neuronal systems. Importantly, these $in$-$silico$ biological findings are consistent and reproducible across three tera-scale virtual mouse brains. Therefore, Tera-MIND showcases a promising path toward efficient and generative simulations of whole organ systems for biomedical research. Project website: https://musikisomorphie.github.io/Tera-MIND.html
SoftCTM: Cell detection by soft instance segmentation and consideration of cell-tissue interaction
Dec 19, 2023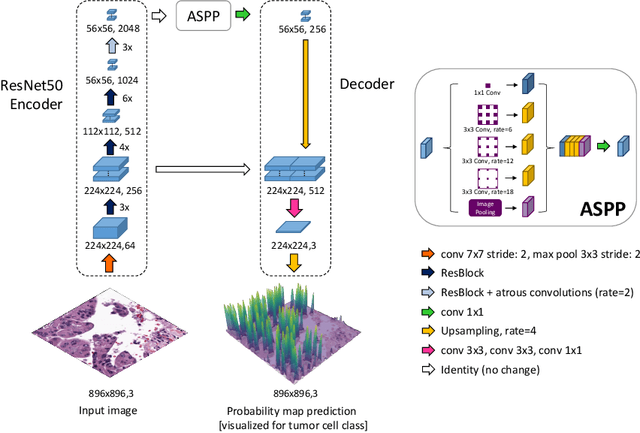
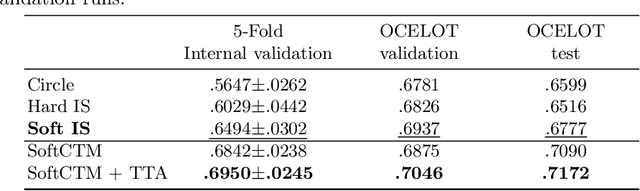
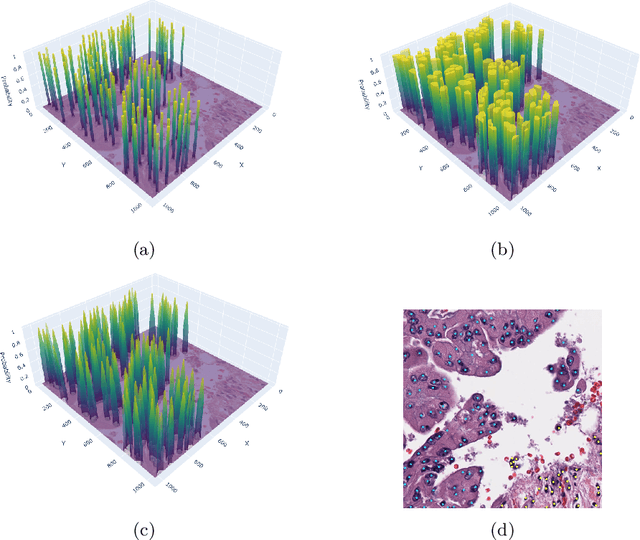

Abstract:Detecting and classifying cells in histopathology H\&E stained whole-slide images is a core task in computational pathology, as it provides valuable insight into the tumor microenvironment. In this work we investigate the impact of ground truth formats on the models performance. Additionally, cell-tissue interactions are considered by providing tissue segmentation predictions as input to the cell detection model. We find that a "soft", probability-map instance segmentation ground truth leads to best model performance. Combined with cell-tissue interaction and test-time augmentation our Soft Cell-Tissue-Model (SoftCTM) achieves 0.7172 mean F1-Score on the Overlapped Cell On Tissue (OCELOT) test set, achieving the third best overall score in the OCELOT 2023 Challenge. The source code for our approach is made publicly available at https://github.com/lely475/ocelot23algo.
Domain generalization across tumor types, laboratories, and species -- insights from the 2022 edition of the Mitosis Domain Generalization Challenge
Sep 27, 2023



Abstract:Recognition of mitotic figures in histologic tumor specimens is highly relevant to patient outcome assessment. This task is challenging for algorithms and human experts alike, with deterioration of algorithmic performance under shifts in image representations. Considerable covariate shifts occur when assessment is performed on different tumor types, images are acquired using different digitization devices, or specimens are produced in different laboratories. This observation motivated the inception of the 2022 challenge on MItosis Domain Generalization (MIDOG 2022). The challenge provided annotated histologic tumor images from six different domains and evaluated the algorithmic approaches for mitotic figure detection provided by nine challenge participants on ten independent domains. Ground truth for mitotic figure detection was established in two ways: a three-expert consensus and an independent, immunohistochemistry-assisted set of labels. This work represents an overview of the challenge tasks, the algorithmic strategies employed by the participants, and potential factors contributing to their success. With an $F_1$ score of 0.764 for the top-performing team, we summarize that domain generalization across various tumor domains is possible with today's deep learning-based recognition pipelines. When assessed against the immunohistochemistry-assisted reference standard, all methods resulted in reduced recall scores, but with only minor changes in the order of participants in the ranking.
CohortFinder: an open-source tool for data-driven partitioning of biomedical image cohorts to yield robust machine learning models
Jul 17, 2023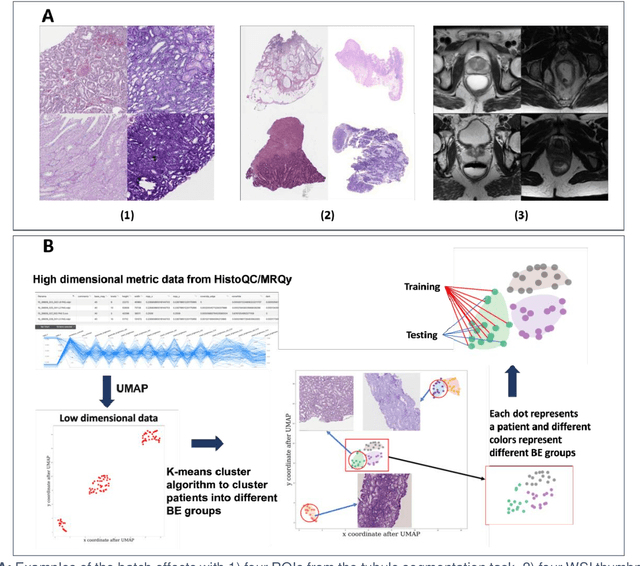

Abstract:Batch effects (BEs) refer to systematic technical differences in data collection unrelated to biological variations whose noise is shown to negatively impact machine learning (ML) model generalizability. Here we release CohortFinder, an open-source tool aimed at mitigating BEs via data-driven cohort partitioning. We demonstrate CohortFinder improves ML model performance in downstream medical image processing tasks. CohortFinder is freely available for download at cohortfinder.com.
Multi-task learning for tissue segmentation and tumor detection in colorectal cancer histology slides
Apr 06, 2023



Abstract:Automating tissue segmentation and tumor detection in histopathology images of colorectal cancer (CRC) is an enabler for faster diagnostic pathology workflows. At the same time it is a challenging task due to low availability of public annotated datasets and high variability of image appearance. The semi-supervised learning for CRC detection (SemiCOL) challenge 2023 provides partially annotated data to encourage the development of automated solutions for tissue segmentation and tumor detection. We propose a U-Net based multi-task model combined with channel-wise and image-statistics-based color augmentations, as well as test-time augmentation, as a candidate solution to the SemiCOL challenge. Our approach achieved a multi-task Dice score of .8655 (Arm 1) and .8515 (Arm 2) for tissue segmentation and AUROC of .9725 (Arm 1) and 0.9750 (Arm 2) for tumor detection on the challenge validation set. The source code for our approach is made publicly available at https://github.com/lely475/CTPLab_SemiCOL2023.
Fine-Grained Hard Negative Mining: Generalizing Mitosis Detection with a Fifth of the MIDOG 2022 Dataset
Jan 03, 2023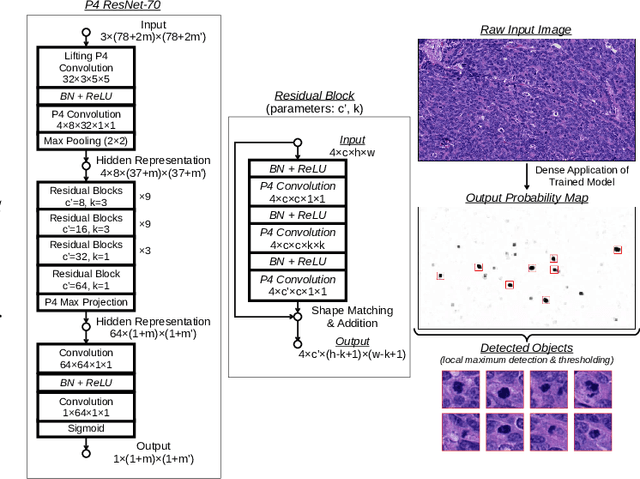
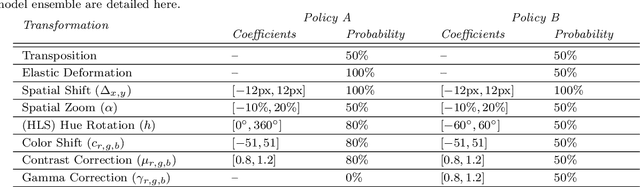
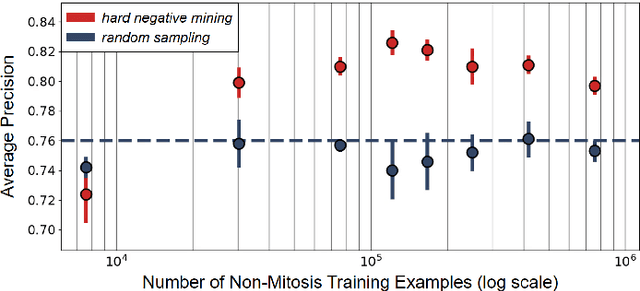
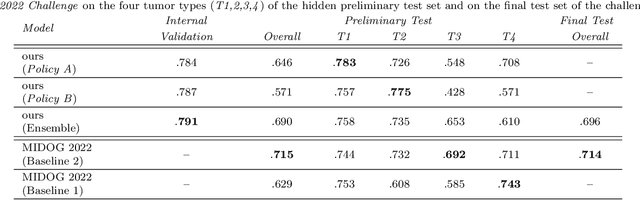
Abstract:Making histopathology image classifiers robust to a wide range of real-world variability is a challenging task. Here, we describe a candidate deep learning solution for the Mitosis Domain Generalization Challenge 2022 (MIDOG) to address the problem of generalization for mitosis detection in images of hematoxylin-eosin-stained histology slides under high variability (scanner, tissue type and species variability). Our approach consists in training a rotation-invariant deep learning model using aggressive data augmentation with a training set enriched with hard negative examples and automatically selected negative examples from the unlabeled part of the challenge dataset. To optimize the performance of our models, we investigated a hard negative mining regime search procedure that lead us to train our best model using a subset of image patches representing 19.6% of our training partition of the challenge dataset. Our candidate model ensemble achieved a F1-score of .697 on the final test set after automated evaluation on the challenge platform, achieving the third best overall score in the MIDOG 2022 Challenge.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022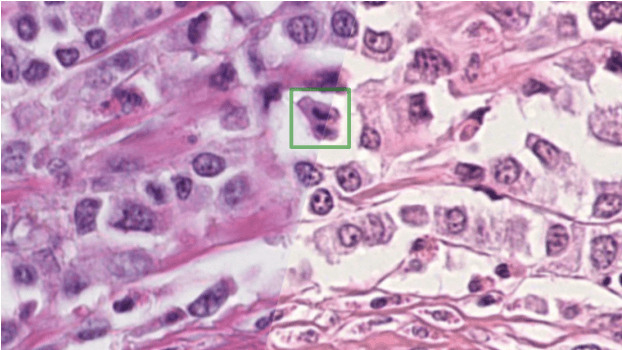
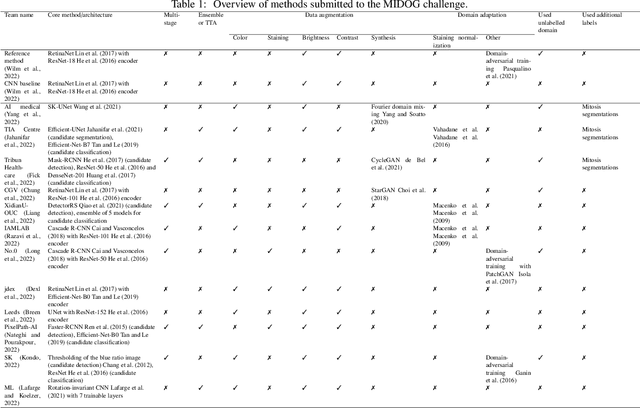
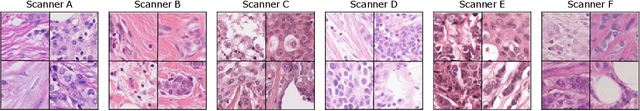
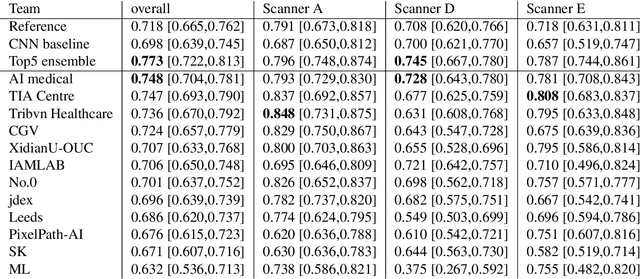
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
Towards IID representation learning and its application on biomedical data
Mar 01, 2022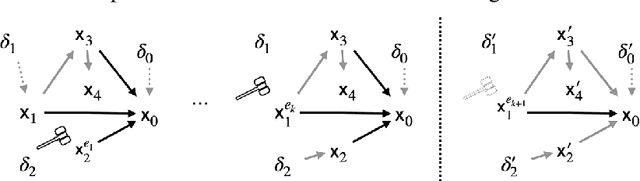
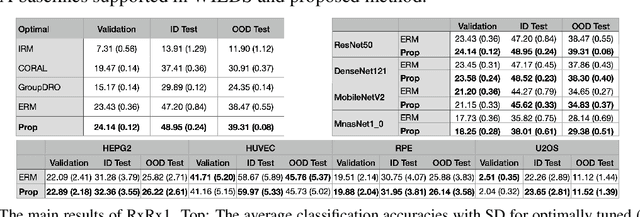
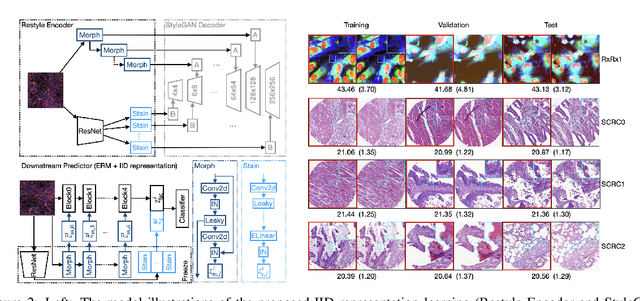
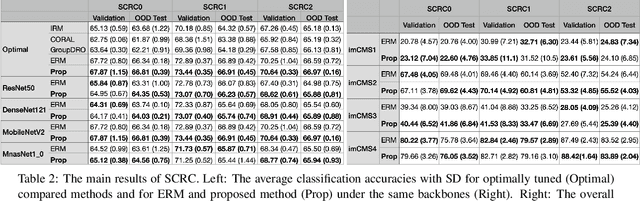
Abstract:Due to the heterogeneity of real-world data, the widely accepted independent and identically distributed (IID) assumption has been criticized in recent studies on causality. In this paper, we argue that instead of being a questionable assumption, IID is a fundamental task-relevant property that needs to be learned. Consider $k$ independent random vectors $\mathsf{X}^{i = 1, \ldots, k}$, we elaborate on how a variety of different causal questions can be reformulated to learning a task-relevant function $\phi$ that induces IID among $\mathsf{Z}^i := \phi \circ \mathsf{X}^i$, which we term IID representation learning. For proof of concept, we examine the IID representation learning on Out-of-Distribution (OOD) generalization tasks. Concretely, by utilizing the representation obtained via the learned function that induces IID, we conduct prediction of molecular characteristics (molecular prediction) on two biomedical datasets with real-world distribution shifts introduced by a) preanalytical variation and b) sampling protocol. To enable reproducibility and for comparison to the state-of-the-art (SOTA) methods, this is done by following the OOD benchmarking guidelines recommended from WILDS. Compared to the SOTA baselines supported in WILDS, the results confirm the superior performance of IID representation learning on OOD tasks. The code is publicly accessible via https://github.com/CTPLab/IID_representation_learning.
Automated causal inference in application to randomized controlled clinical trials
Jan 20, 2022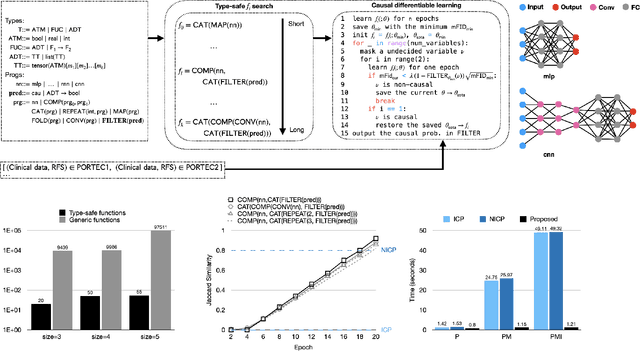
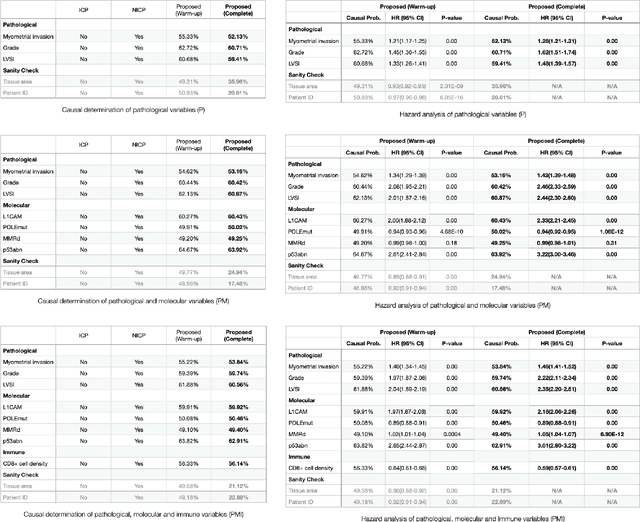
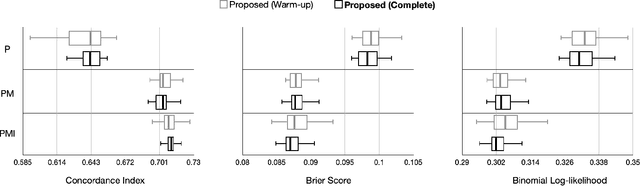
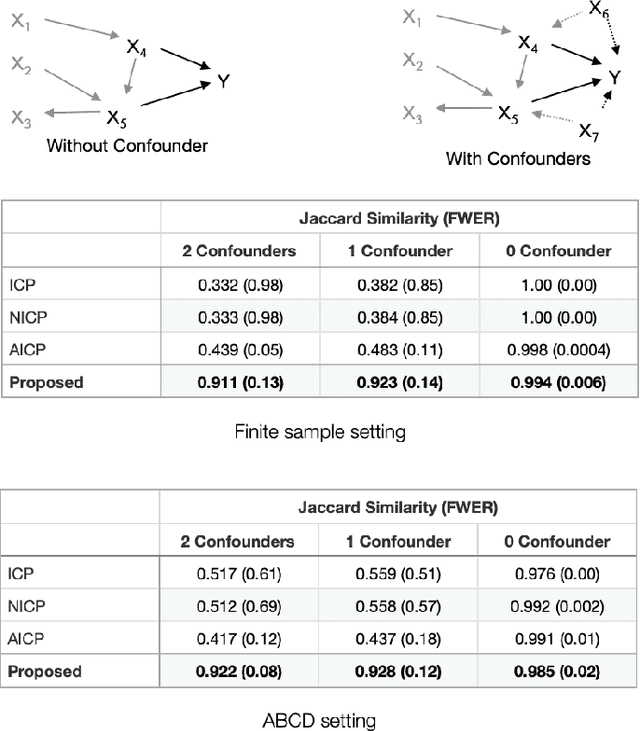
Abstract:Randomized controlled trials (RCTs) are considered as the gold standard for testing causal hypotheses in the clinical domain. However, the investigation of prognostic variables of patient outcome in a hypothesized cause-effect route is not feasible using standard statistical methods. Here, we propose a new automated causal inference method (AutoCI) built upon the invariant causal prediction (ICP) framework for the causal re-interpretation of clinical trial data. Compared to existing methods, we show that the proposed AutoCI allows to efficiently determine the causal variables with a clear differentiation on two real-world RCTs of endometrial cancer patients with mature outcome and extensive clinicopathological and molecular data. This is achieved via suppressing the causal probability of non-causal variables by a wide margin. In ablation studies, we further demonstrate that the assignment of causal probabilities by AutoCI remain consistent in the presence of confounders. In conclusion, these results confirm the robustness and feasibility of AutoCI for future applications in real-world clinical analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge