Inti Zlobec
A CAD System for Colorectal Cancer from WSI: A Clinically Validated Interpretable ML-based Prototype
Jan 06, 2023Abstract:The integration of Artificial Intelligence (AI) and Digital Pathology has been increasing over the past years. Nowadays, applications of deep learning (DL) methods to diagnose cancer from whole-slide images (WSI) are, more than ever, a reality within different research groups. Nonetheless, the development of these systems was limited by a myriad of constraints regarding the lack of training samples, the scaling difficulties, the opaqueness of DL methods, and, more importantly, the lack of clinical validation. As such, we propose a system designed specifically for the diagnosis of colorectal samples. The construction of such a system consisted of four stages: (1) a careful data collection and annotation process, which resulted in one of the largest WSI colorectal samples datasets; (2) the design of an interpretable mixed-supervision scheme to leverage the domain knowledge introduced by pathologists through spatial annotations; (3) the development of an effective sampling approach based on the expected severeness of each tile, which decreased the computation cost by a factor of almost 6x; (4) the creation of a prototype that integrates the full set of features of the model to be evaluated in clinical practice. During these stages, the proposed method was evaluated in four separate test sets, two of them are external and completely independent. On the largest of those sets, the proposed approach achieved an accuracy of 93.44%. DL for colorectal samples is a few steps closer to stop being research exclusive and to become fully integrated in clinical practice.
Panoptic segmentation with highly imbalanced semantic labels
Apr 02, 2022



Abstract:This manuscript describes the panoptic segmentation method we devised for our submission to the CONIC challenge at ISBI 2022. Key features of our method are a weighted loss that we specifically engineered for semantic segmentation of highly imbalanced cell types, and an existing state-of-the art nuclei instance segmentation model, which we combine in a Hovernet-like architecture.
Towards IID representation learning and its application on biomedical data
Mar 01, 2022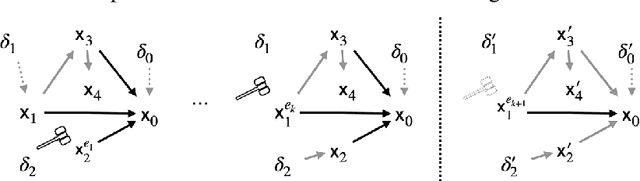
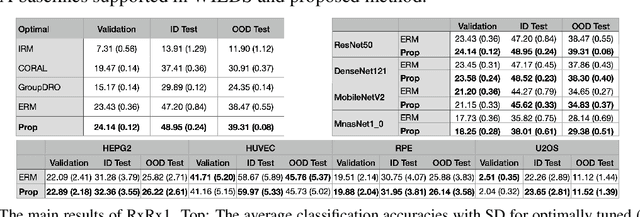
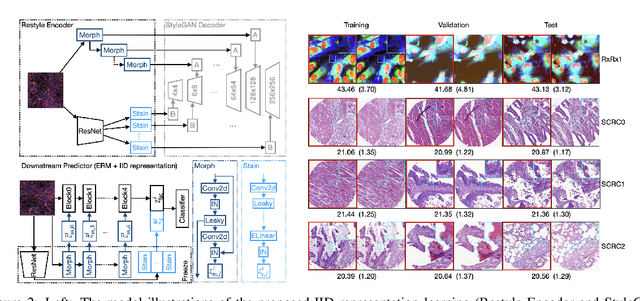
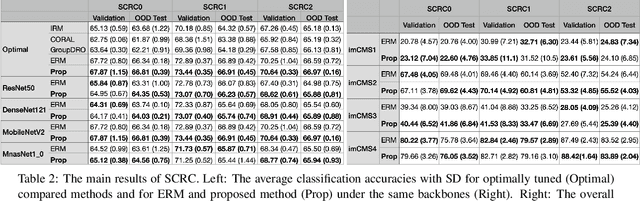
Abstract:Due to the heterogeneity of real-world data, the widely accepted independent and identically distributed (IID) assumption has been criticized in recent studies on causality. In this paper, we argue that instead of being a questionable assumption, IID is a fundamental task-relevant property that needs to be learned. Consider $k$ independent random vectors $\mathsf{X}^{i = 1, \ldots, k}$, we elaborate on how a variety of different causal questions can be reformulated to learning a task-relevant function $\phi$ that induces IID among $\mathsf{Z}^i := \phi \circ \mathsf{X}^i$, which we term IID representation learning. For proof of concept, we examine the IID representation learning on Out-of-Distribution (OOD) generalization tasks. Concretely, by utilizing the representation obtained via the learned function that induces IID, we conduct prediction of molecular characteristics (molecular prediction) on two biomedical datasets with real-world distribution shifts introduced by a) preanalytical variation and b) sampling protocol. To enable reproducibility and for comparison to the state-of-the-art (SOTA) methods, this is done by following the OOD benchmarking guidelines recommended from WILDS. Compared to the SOTA baselines supported in WILDS, the results confirm the superior performance of IID representation learning on OOD tasks. The code is publicly accessible via https://github.com/CTPLab/IID_representation_learning.
Self-Rule to Adapt: Generalized Multi-source Feature Learning Using Unsupervised Domain Adaptation for Colorectal Cancer Tissue Detection
Aug 20, 2021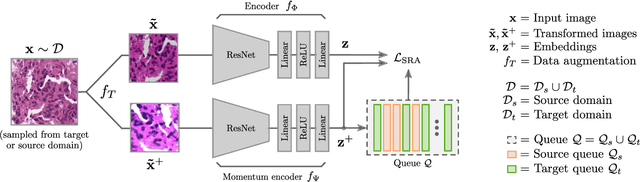
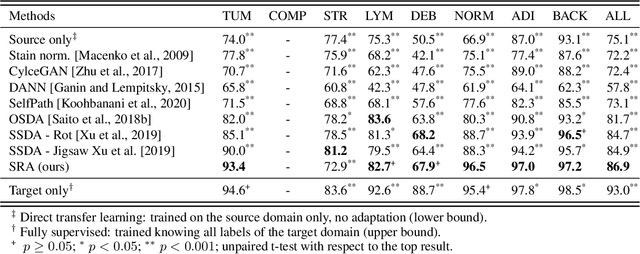
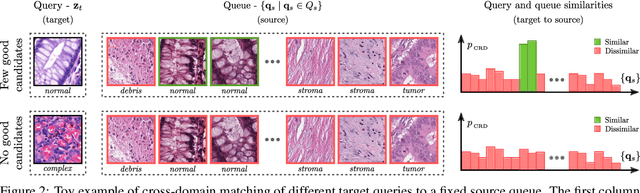
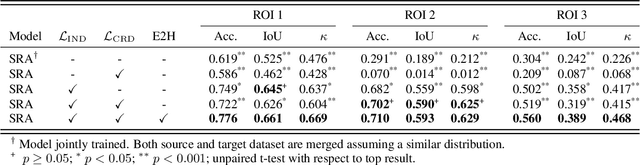
Abstract:Supervised learning is constrained by the availability of labeled data, which are especially expensive to acquire in the field of digital pathology. Making use of open-source data for pre-training or using domain adaptation can be a way to overcome this issue. However, pre-trained networks often fail to generalize to new test domains that are not distributed identically due to variations in tissue stainings, types, and textures. Additionally, current domain adaptation methods mainly rely on fully-labeled source datasets. In this work, we propose SRA, which takes advantage of self-supervised learning to perform domain adaptation and removes the necessity of a fully-labeled source dataset. SRA can effectively transfer the discriminative knowledge obtained from a few labeled source domain's data to a new target domain without requiring additional tissue annotations. Our method harnesses both domains' structures by capturing visual similarity with intra-domain and cross-domain self-supervision. Moreover, we present a generalized formulation of our approach that allows the architecture to learn from multi-source domains. We show that our proposed method outperforms baselines for domain adaptation of colorectal tissue type classification and further validate our approach on our in-house clinical cohort. The code and models are available open-source: https://github.com/christianabbet/SRA.
Divide-and-Rule: Self-Supervised Learning for Survival Analysis in Colorectal Cancer
Jul 07, 2020



Abstract:With the long-term rapid increase in incidences of colorectal cancer (CRC), there is an urgent clinical need to improve risk stratification. The conventional pathology report is usually limited to only a few histopathological features. However, most of the tumor microenvironments used to describe patterns of aggressive tumor behavior are ignored. In this work, we aim to learn histopathological patterns within cancerous tissue regions that can be used to improve prognostic stratification for colorectal cancer. To do so, we propose a self-supervised learning method that jointly learns a representation of tissue regions as well as a metric of the clustering to obtain their underlying patterns. These histopathological patterns are then used to represent the interaction between complex tissues and predict clinical outcomes directly. We furthermore show that the proposed approach can benefit from linear predictors to avoid overfitting in patient outcomes predictions. To this end, we introduce a new well-characterized clinicopathological dataset, including a retrospective collective of 374 patients, with their survival time and treatment information. Histomorphological clusters obtained by our method are evaluated by training survival models. The experimental results demonstrate statistically significant patient stratification, and our approach outperformed the state-of-the-art deep clustering methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge