Peter Hirsch
FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures
Mar 29, 2024



Abstract:Instance segmentation of neurons in volumetric light microscopy images of nervous systems enables groundbreaking research in neuroscience by facilitating joint functional and morphological analyses of neural circuits at cellular resolution. Yet said multi-neuron light microscopy data exhibits extremely challenging properties for the task of instance segmentation: Individual neurons have long-ranging, thin filamentous and widely branching morphologies, multiple neurons are tightly inter-weaved, and partial volume effects, uneven illumination and noise inherent to light microscopy severely impede local disentangling as well as long-range tracing of individual neurons. These properties reflect a current key challenge in machine learning research, namely to effectively capture long-range dependencies in the data. While respective methodological research is buzzing, to date methods are typically benchmarked on synthetic datasets. To address this gap, we release the FlyLight Instance Segmentation Benchmark (FISBe) dataset, the first publicly available multi-neuron light microscopy dataset with pixel-wise annotations. In addition, we define a set of instance segmentation metrics for benchmarking that we designed to be meaningful with regard to downstream analyses. Lastly, we provide three baselines to kick off a competition that we envision to both advance the field of machine learning regarding methodology for capturing long-range data dependencies, and facilitate scientific discovery in basic neuroscience.
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
Tracking by weakly-supervised learning and graph optimization for whole-embryo C. elegans lineages
Aug 24, 2022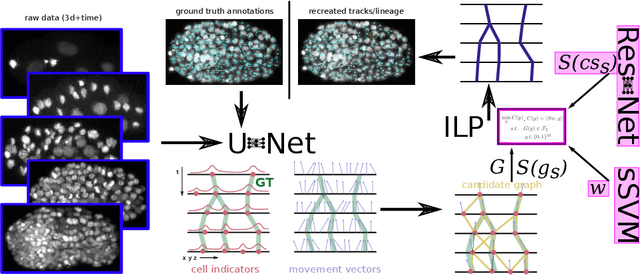
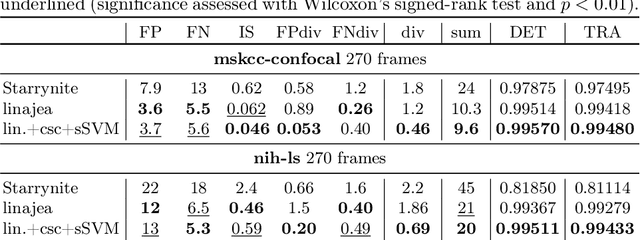
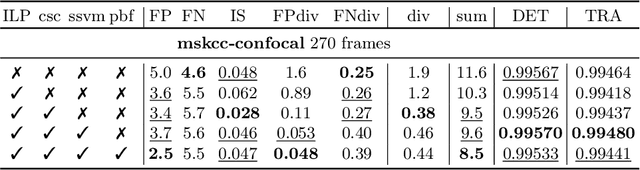
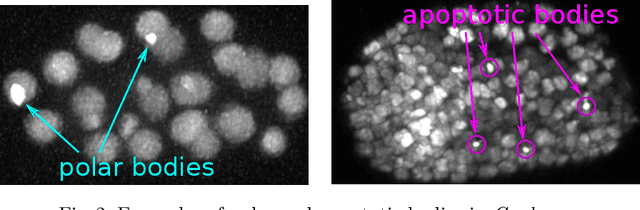
Abstract:Tracking all nuclei of an embryo in noisy and dense fluorescence microscopy data is a challenging task. We build upon a recent method for nuclei tracking that combines weakly-supervised learning from a small set of nuclei center point annotations with an integer linear program (ILP) for optimal cell lineage extraction. Our work specifically addresses the following challenging properties of C. elegans embryo recordings: (1) Many cell divisions as compared to benchmark recordings of other organisms, and (2) the presence of polar bodies that are easily mistaken as cell nuclei. To cope with (1), we devise and incorporate a learnt cell division detector. To cope with (2), we employ a learnt polar body detector. We further propose automated ILP weights tuning via a structured SVM, alleviating the need for tedious manual set-up of a respective grid search. Our method outperforms the previous leader of the cell tracking challenge on the Fluo-N3DH-CE embryo dataset. We report a further extensive quantitative evaluation on two more C. elegans datasets. We will make these datasets public to serve as an extended benchmark for future method development. Our results suggest considerable improvements yielded by our method, especially in terms of the correctness of division event detection and the number and length of fully correct track segments. Code: https://github.com/funkelab/linajea
Panoptic segmentation with highly imbalanced semantic labels
Apr 02, 2022



Abstract:This manuscript describes the panoptic segmentation method we devised for our submission to the CONIC challenge at ISBI 2022. Key features of our method are a weighted loss that we specifically engineered for semantic segmentation of highly imbalanced cell types, and an existing state-of-the art nuclei instance segmentation model, which we combine in a Hovernet-like architecture.
How Shift Equivariance Impacts Metric Learning for Instance Segmentation
Jan 14, 2021



Abstract:Metric learning has received conflicting assessments concerning its suitability for solving instance segmentation tasks. It has been dismissed as theoretically flawed due to the shift equivariance of the employed CNNs and their respective inability to distinguish same-looking objects. Yet it has been shown to yield state of the art results for a variety of tasks, and practical issues have mainly been reported in the context of tile-and-stitch approaches, where discontinuities at tile boundaries have been observed. To date, neither of the reported issues have undergone thorough formal analysis. In our work, we contribute a comprehensive formal analysis of the shift equivariance properties of encoder-decoder-style CNNs, which yields a clear picture of what can and cannot be achieved with metric learning in the face of same-looking objects. In particular, we prove that a standard encoder-decoder network that takes $d$-dimensional images as input, with $l$ pooling layers and pooling factor $f$, has the capacity to distinguish at most $f^{dl}$ same-looking objects, and we show that this upper limit can be reached. Furthermore, we show that to avoid discontinuities in a tile-and-stitch approach, assuming standard batch size 1, it is necessary to employ valid convolutions in combination with a training output window size strictly greater than $f^l$, while at test-time it is necessary to crop tiles to size $n\cdot f^l$ before stitching, with $n\geq 1$. We complement these theoretical findings by discussing a number of insightful special cases for which we show empirical results on synthetic data.
An Auxiliary Task for Learning Nuclei Segmentation in 3D Microscopy Images
Feb 07, 2020



Abstract:Segmentation of cell nuclei in microscopy images is a prevalent necessity in cell biology. Especially for three-dimensional datasets, manual segmentation is prohibitively time-consuming, motivating the need for automated methods. Learning-based methods trained on pixel-wise ground-truth segmentations have been shown to yield state-of-the-art results on 2d benchmark image data of nuclei, yet a respective benchmark is missing for 3d image data. In this work, we perform a comparative evaluation of nuclei segmentation algorithms on a database of manually segmented 3d light microscopy volumes. We propose a novel learning strategy that boosts segmentation accuracy by means of a simple auxiliary task, thereby robustly outperforming each of our baselines. Furthermore, we show that one of our baselines, the popular three-label model, when trained with our proposed auxiliary task, outperforms the recent StarDist-3D. As an additional, practical contribution, we benchmark nuclei segmentation against nuclei detection, i.e. the task of merely pinpointing individual nuclei without generating respective pixel-accurate segmentations. For learning nuclei detection, large 3d training datasets of manually annotated nuclei center points are available. However, the impact on detection accuracy caused by training on such sparse ground truth as opposed to dense pixel-wise ground truth has not yet been quantified. To this end, we compare nuclei detection accuracy yielded by training on dense vs. sparse ground truth. Our results suggest that training on sparse ground truth yields competitive nuclei detection rates.
PatchPerPix for Instance Segmentation
Jan 21, 2020



Abstract:In this paper we present a novel method for proposal free instance segmentation that can handle sophisticated object shapes that span large parts of an image and form dense object clusters with crossovers. Our method is based on predicting dense local shape descriptors, which we assemble to form instances. All instances are assembled simultaneously in one go. To our knowledge, our method is the first non-iterative method that guarantees instances to be composed of learnt shape patches. We evaluate our method on a variety of data domains, where it defines the new state of the art on two challenging benchmarks, namely the ISBI 2012 EM segmentation benchmark, and the BBBC010 C. elegans dataset. We show furthermore that our method performs well also on 3d image data, and can handle even extreme cases of complex shape clusters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge