Caroline Malin-Mayor
DaCapo: a modular deep learning framework for scalable 3D image segmentation
Aug 05, 2024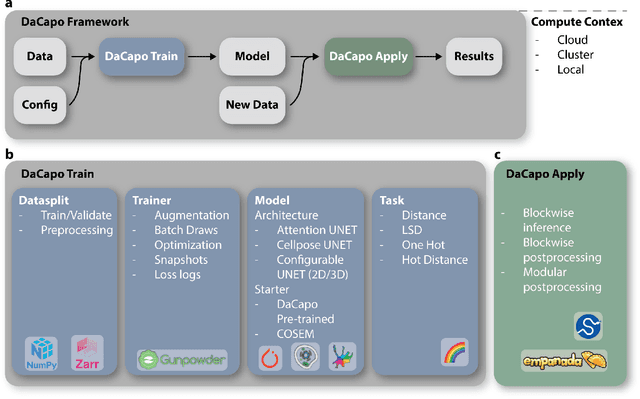
Abstract:DaCapo is a specialized deep learning library tailored to expedite the training and application of existing machine learning approaches on large, near-isotropic image data. In this correspondence, we introduce DaCapo's unique features optimized for this specific domain, highlighting its modular structure, efficient experiment management tools, and scalable deployment capabilities. We discuss its potential to improve access to large-scale, isotropic image segmentation and invite the community to explore and contribute to this open-source initiative.
Pose2Gait: Extracting Gait Features from Monocular Video of Individuals with Dementia
Aug 22, 2023



Abstract:Video-based ambient monitoring of gait for older adults with dementia has the potential to detect negative changes in health and allow clinicians and caregivers to intervene early to prevent falls or hospitalizations. Computer vision-based pose tracking models can process video data automatically and extract joint locations; however, publicly available models are not optimized for gait analysis on older adults or clinical populations. In this work we train a deep neural network to map from a two dimensional pose sequence, extracted from a video of an individual walking down a hallway toward a wall-mounted camera, to a set of three-dimensional spatiotemporal gait features averaged over the walking sequence. The data of individuals with dementia used in this work was captured at two sites using a wall-mounted system to collect the video and depth information used to train and evaluate our model. Our Pose2Gait model is able to extract velocity and step length values from the video that are correlated with the features from the depth camera, with Spearman's correlation coefficients of .83 and .60 respectively, showing that three dimensional spatiotemporal features can be predicted from monocular video. Future work remains to improve the accuracy of other features, such as step time and step width, and test the utility of the predicted values for detecting meaningful changes in gait during longitudinal ambient monitoring.
Tracking by weakly-supervised learning and graph optimization for whole-embryo C. elegans lineages
Aug 24, 2022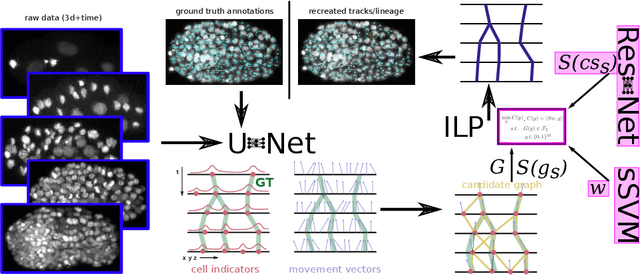
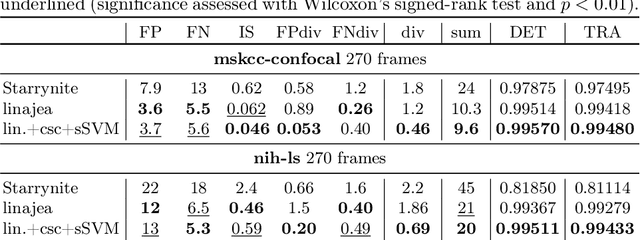
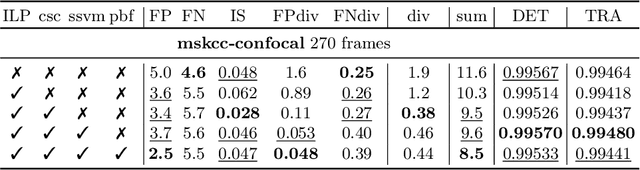
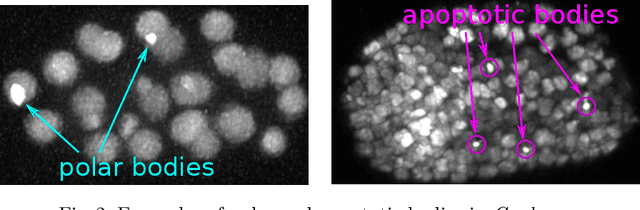
Abstract:Tracking all nuclei of an embryo in noisy and dense fluorescence microscopy data is a challenging task. We build upon a recent method for nuclei tracking that combines weakly-supervised learning from a small set of nuclei center point annotations with an integer linear program (ILP) for optimal cell lineage extraction. Our work specifically addresses the following challenging properties of C. elegans embryo recordings: (1) Many cell divisions as compared to benchmark recordings of other organisms, and (2) the presence of polar bodies that are easily mistaken as cell nuclei. To cope with (1), we devise and incorporate a learnt cell division detector. To cope with (2), we employ a learnt polar body detector. We further propose automated ILP weights tuning via a structured SVM, alleviating the need for tedious manual set-up of a respective grid search. Our method outperforms the previous leader of the cell tracking challenge on the Fluo-N3DH-CE embryo dataset. We report a further extensive quantitative evaluation on two more C. elegans datasets. We will make these datasets public to serve as an extended benchmark for future method development. Our results suggest considerable improvements yielded by our method, especially in terms of the correctness of division event detection and the number and length of fully correct track segments. Code: https://github.com/funkelab/linajea
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge