Samir Jabari
On the Value of PHH3 for Mitotic Figure Detection on H&E-stained Images
Jun 28, 2024Abstract:The count of mitotic figures (MFs) observed in hematoxylin and eosin (H&E)-stained slides is an important prognostic marker as it is a measure for tumor cell proliferation. However, the identification of MFs has a known low inter-rater agreement. Deep learning algorithms can standardize this task, but they require large amounts of annotated data for training and validation. Furthermore, label noise introduced during the annotation process may impede the algorithm's performance. Unlike H&E, the mitosis-specific antibody phospho-histone H3 (PHH3) specifically highlights MFs. Counting MFs on slides stained against PHH3 leads to higher agreement among raters and has therefore recently been used as a ground truth for the annotation of MFs in H&E. However, as PHH3 facilitates the recognition of cells indistinguishable from H&E stain alone, the use of this ground truth could potentially introduce noise into the H&E-related dataset, impacting model performance. This study analyzes the impact of PHH3-assisted MF annotation on inter-rater reliability and object level agreement through an extensive multi-rater experiment. We found that the annotators' object-level agreement increased when using PHH3-assisted labeling. Subsequently, MF detectors were evaluated on the resulting datasets to investigate the influence of PHH3-assisted labeling on the models' performance. Additionally, a novel dual-stain MF detector was developed to investigate the interpretation-shift of PHH3-assisted labels used in H&E, which clearly outperformed single-stain detectors. However, the PHH3-assisted labels did not have a positive effect on solely H&E-based models. The high performance of our dual-input detector reveals an information mismatch between the H&E and PHH3-stained images as the cause of this effect.
Re-identification from histopathology images
Mar 19, 2024



Abstract:In numerous studies, deep learning algorithms have proven their potential for the analysis of histopathology images, for example, for revealing the subtypes of tumors or the primary origin of metastases. These models require large datasets for training, which must be anonymized to prevent possible patient identity leaks. This study demonstrates that even relatively simple deep learning algorithms can re-identify patients in large histopathology datasets with substantial accuracy. We evaluated our algorithms on two TCIA datasets including lung squamous cell carcinoma (LSCC) and lung adenocarcinoma (LUAD). We also demonstrate the algorithm's performance on an in-house dataset of meningioma tissue. We predicted the source patient of a slide with F1 scores of 50.16 % and 52.30 % on the LSCC and LUAD datasets, respectively, and with 62.31 % on our meningioma dataset. Based on our findings, we formulated a risk assessment scheme to estimate the risk to the patient's privacy prior to publication.
Domain generalization across tumor types, laboratories, and species -- insights from the 2022 edition of the Mitosis Domain Generalization Challenge
Sep 27, 2023



Abstract:Recognition of mitotic figures in histologic tumor specimens is highly relevant to patient outcome assessment. This task is challenging for algorithms and human experts alike, with deterioration of algorithmic performance under shifts in image representations. Considerable covariate shifts occur when assessment is performed on different tumor types, images are acquired using different digitization devices, or specimens are produced in different laboratories. This observation motivated the inception of the 2022 challenge on MItosis Domain Generalization (MIDOG 2022). The challenge provided annotated histologic tumor images from six different domains and evaluated the algorithmic approaches for mitotic figure detection provided by nine challenge participants on ten independent domains. Ground truth for mitotic figure detection was established in two ways: a three-expert consensus and an independent, immunohistochemistry-assisted set of labels. This work represents an overview of the challenge tasks, the algorithmic strategies employed by the participants, and potential factors contributing to their success. With an $F_1$ score of 0.764 for the top-performing team, we summarize that domain generalization across various tumor domains is possible with today's deep learning-based recognition pipelines. When assessed against the immunohistochemistry-assisted reference standard, all methods resulted in reduced recall scores, but with only minor changes in the order of participants in the ranking.
SYNTA: A novel approach for deep learning-based image analysis in muscle histopathology using photo-realistic synthetic data
Aug 03, 2022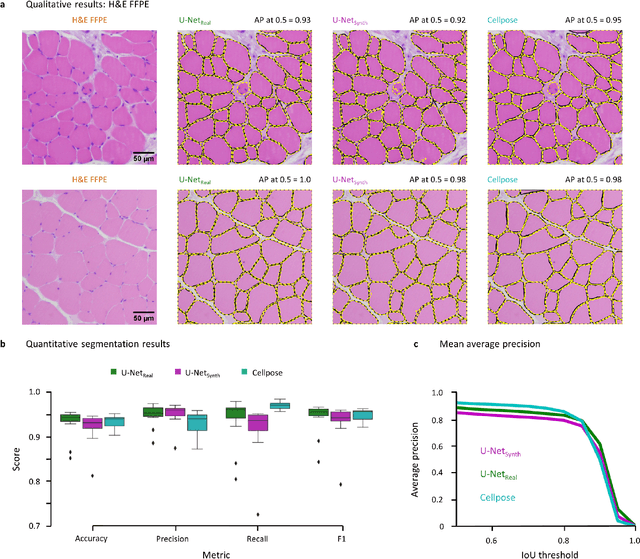
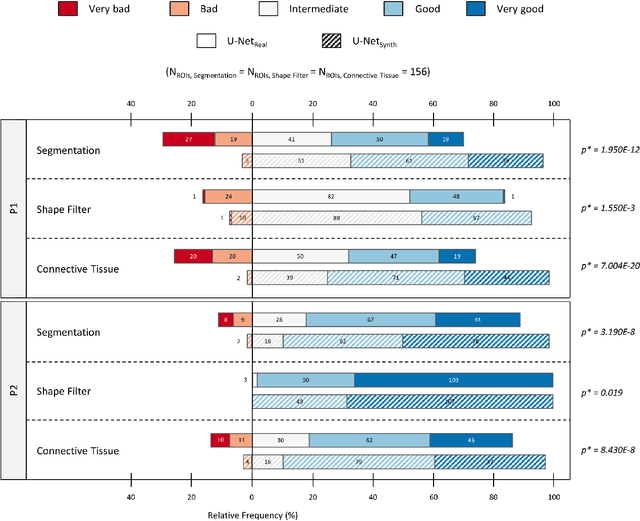
Abstract:Artificial intelligence (AI), machine learning, and deep learning (DL) methods are becoming increasingly important in the field of biomedical image analysis. However, to exploit the full potential of such methods, a representative number of experimentally acquired images containing a significant number of manually annotated objects is needed as training data. Here we introduce SYNTA (synthetic data) as a novel approach for the generation of synthetic, photo-realistic, and highly complex biomedical images as training data for DL systems. We show the versatility of our approach in the context of muscle fiber and connective tissue analysis in histological sections. We demonstrate that it is possible to perform robust and expert-level segmentation tasks on previously unseen real-world data, without the need for manual annotations using synthetic training data alone. Being a fully parametric technique, our approach poses an interpretable and controllable alternative to Generative Adversarial Networks (GANs) and has the potential to significantly accelerate quantitative image analysis in a variety of biomedical applications in microscopy and beyond.
Automatic and explainable grading of meningiomas from histopathology images
Jul 19, 2021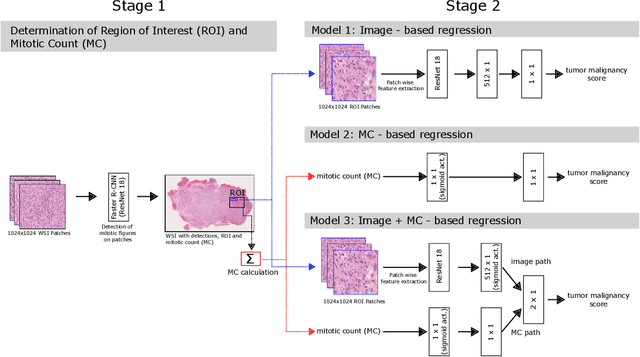
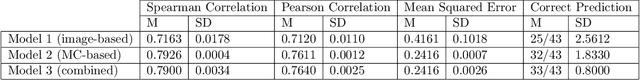
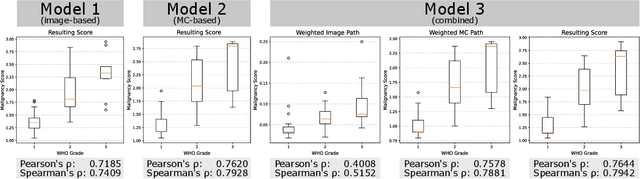
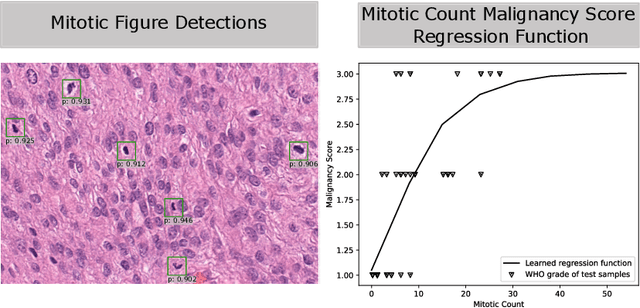
Abstract:Meningioma is one of the most prevalent brain tumors in adults. To determine its malignancy, it is graded by a pathologist into three grades according to WHO standards. This grade plays a decisive role in treatment, and yet may be subject to inter-rater discordance. In this work, we present and compare three approaches towards fully automatic meningioma grading from histology whole slide images. All approaches are following a two-stage paradigm, where we first identify a region of interest based on the detection of mitotic figures in the slide using a state-of-the-art object detection deep learning network. This region of highest mitotic rate is considered characteristic for biological tumor behavior. In the second stage, we calculate a score corresponding to tumor malignancy based on information contained in this region using three different settings. In a first approach, image patches are sampled from this region and regression is based on morphological features encoded by a ResNet-based network. We compare this to learning a logistic regression from the determined mitotic count, an approach which is easily traceable and explainable. Lastly, we combine both approaches in a single network. We trained the pipeline on 951 slides from 341 patients and evaluated them on a separate set of 141 slides from 43 patients. All approaches yield a high correlation to the WHO grade. The logistic regression and the combined approach had the best results in our experiments, yielding correct predictions in 32 and 33 of all cases, respectively, with the image-based approach only predicting 25 cases correctly. Spearman's correlation was 0.716, 0.792 and 0.790 respectively. It may seem counterintuitive at first that morphological features provided by image patches do not improve model performance. Yet, this mirrors the criteria of the grading scheme, where mitotic count is the only unequivocal parameter.
Learning New Tricks from Old Dogs -- Inter-Species, Inter-Tissue Domain Adaptation for Mitotic Figure Assessment
Nov 25, 2019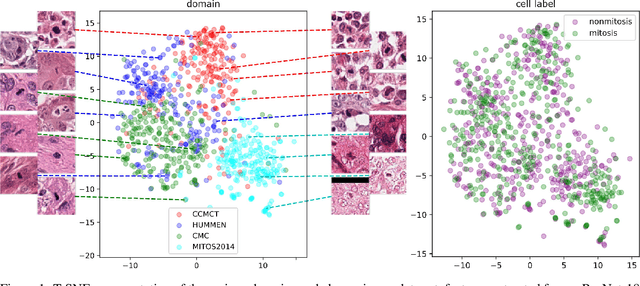
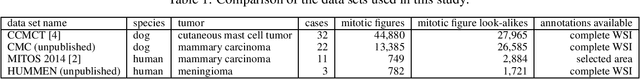
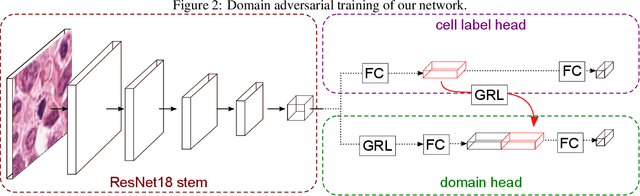
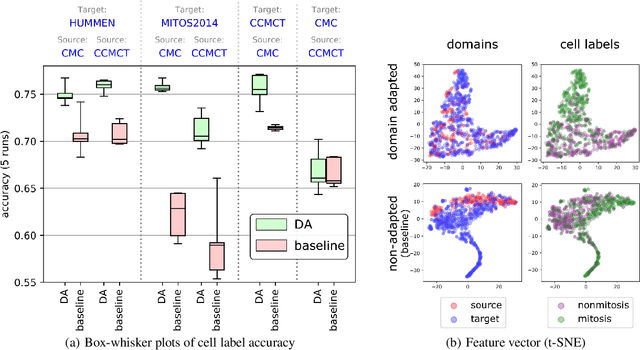
Abstract:For histopathological tumor assessment, the count of mitotic figures per area is an important part of prognostication. Algorithmic approaches - such as for mitotic figure identification - have significantly improved in recent times, potentially allowing for computer-augmented or fully automatic screening systems in the future. This trend is further supported by whole slide scanning microscopes becoming available in many pathology labs and could soon become a standard imaging tool. For an application in broader fields of such algorithms, the availability of mitotic figure data sets of sufficient size for the respective tissue type and species is an important precondition, that is, however, rarely met. While algorithmic performance climbed steadily for e.g. human mammary carcinoma, thanks to several challenges held in the field, for most tumor types, data sets are not available. In this work, we assess domain transfer of mitotic figure recognition using domain adversarial training on four data sets, two from dogs and two from humans. We were able to show that domain adversarial training considerably improves accuracy when applying mitotic figure classification learned from the canine on the human data sets (up to +12.8% in accuracy) and is thus a helpful method to transfer knowledge from existing data sets to new tissue types and species.
Deep Learning-Based Quantification of Pulmonary Hemosiderophages in Cytology Slides
Aug 12, 2019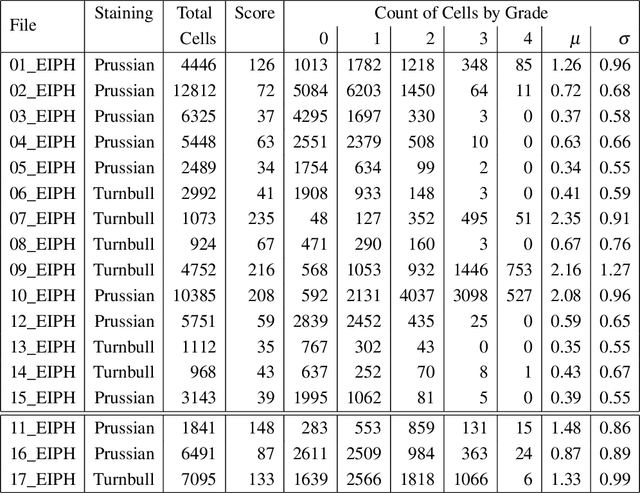
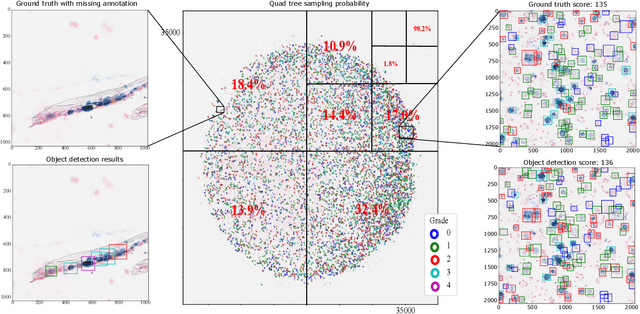
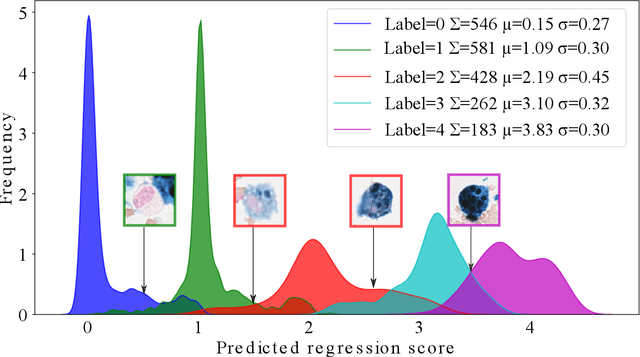
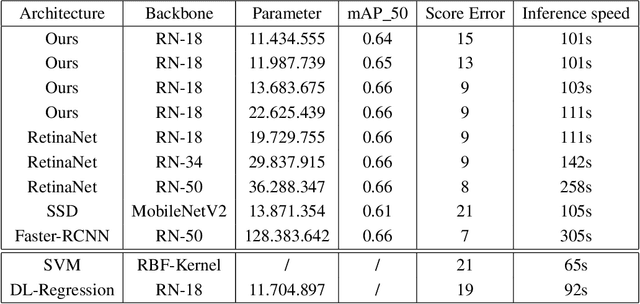
Abstract:Purpose: Exercise-induced pulmonary hemorrhage (EIPH) is a common syndrome in sport horses with negative impact on performance. Cytology of bronchoalveolar lavage fluid by use of a scoring system is considered the most sensitive diagnostic method. Macrophages are classified depending on the degree of cytoplasmic hemosiderin content. The current gold standard is manual grading, which is however monotonous and time-consuming. Methods: We evaluated state-of-the-art deep learning-based methods for single cell macrophage classification and compared them against the performance of nine cytology experts and evaluated inter- and intra-observer variability. Additionally, we evaluated object detection methods on a novel data set of 17 completely annotated cytology whole slide images (WSI) containing 78,047 hemosiderophages. Resultsf: Our deep learning-based approach reached a concordance of 0.85, partially exceeding human expert concordance (0.68 to 0.86, $\mu$=0.73, $\sigma$ =0.04). Intra-observer variability was high (0.68 to 0.88) and inter-observer concordance was moderate (Fleiss kappa = 0.67). Our object detection approach has a mean average precision of 0.66 over the five classes from the whole slide gigapixel image and a computation time of below two minutes. Conclusion: To mitigate the high inter- and intra-rater variability, we propose our automated object detection pipeline, enabling accurate, reproducible and quick EIPH scoring in WSI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge