Georg Schett
ShaRPy: Shape Reconstruction and Hand Pose Estimation from RGB-D with Uncertainty
Mar 17, 2023Abstract:Despite their potential, markerless hand tracking technologies are not yet applied in practice to the diagnosis or monitoring of the activity in inflammatory musculoskeletal diseases. One reason is that the focus of most methods lies in the reconstruction of coarse, plausible poses for gesture recognition or AR/VR applications, whereas in the clinical context, accurate, interpretable, and reliable results are required. Therefore, we propose ShaRPy, the first RGB-D Shape Reconstruction and hand Pose tracking system, which provides uncertainty estimates of the computed pose to guide clinical decision-making. Our method requires only a light-weight setup with a single consumer-level RGB-D camera yet it is able to distinguish similar poses with only small joint angle deviations. This is achieved by combining a data-driven dense correspondence predictor with traditional energy minimization, optimizing for both, pose and hand shape parameters. We evaluate ShaRPy on a keypoint detection benchmark and show qualitative results on recordings of a patient.
Motion Compensation via Epipolar Consistency for In-Vivo X-Ray Microscopy
Mar 01, 2023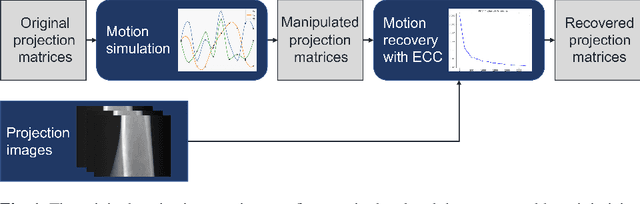
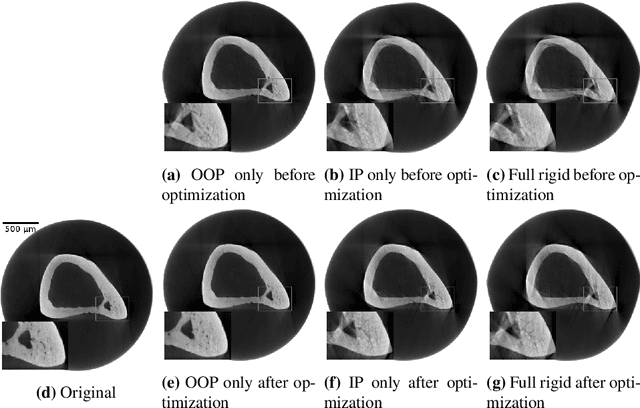
Abstract:Intravital X-ray microscopy (XRM) in preclinical mouse models is of vital importance for the identification of microscopic structural pathological changes in the bone which are characteristic of osteoporosis. The complexity of this method stems from the requirement for high-quality 3D reconstructions of the murine bones. However, respiratory motion and muscle relaxation lead to inconsistencies in the projection data which result in artifacts in uncompensated reconstructions. Motion compensation using epipolar consistency conditions (ECC) has previously shown good performance in clinical CT settings. Here, we explore whether such algorithms are suitable for correcting motion-corrupted XRM data. Different rigid motion patterns are simulated and the quality of the motion-compensated reconstructions is assessed. The method is able to restore microscopic features for out-of-plane motion, but artifacts remain for more realistic motion patterns including all six degrees of freedom of rigid motion. Therefore, ECC is valuable for the initial alignment of the projection data followed by further fine-tuning of motion parameters using a reconstruction-based method
SYNTA: A novel approach for deep learning-based image analysis in muscle histopathology using photo-realistic synthetic data
Aug 03, 2022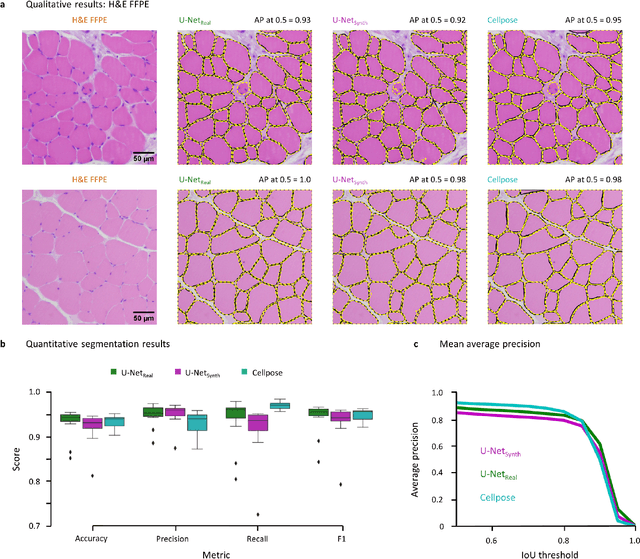
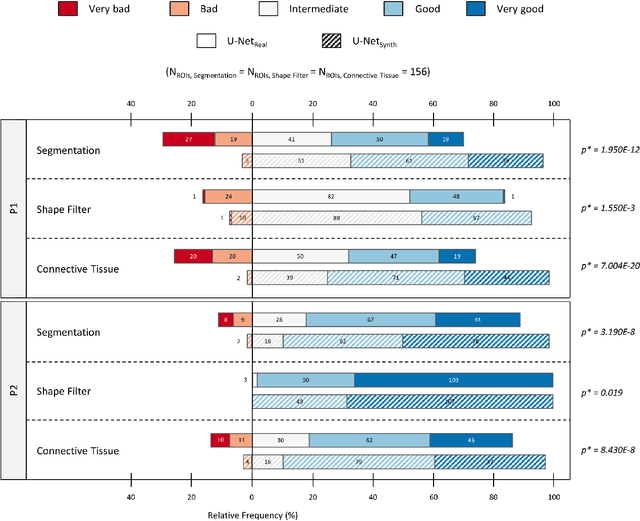
Abstract:Artificial intelligence (AI), machine learning, and deep learning (DL) methods are becoming increasingly important in the field of biomedical image analysis. However, to exploit the full potential of such methods, a representative number of experimentally acquired images containing a significant number of manually annotated objects is needed as training data. Here we introduce SYNTA (synthetic data) as a novel approach for the generation of synthetic, photo-realistic, and highly complex biomedical images as training data for DL systems. We show the versatility of our approach in the context of muscle fiber and connective tissue analysis in histological sections. We demonstrate that it is possible to perform robust and expert-level segmentation tasks on previously unseen real-world data, without the need for manual annotations using synthetic training data alone. Being a fully parametric technique, our approach poses an interpretable and controllable alternative to Generative Adversarial Networks (GANs) and has the potential to significantly accelerate quantitative image analysis in a variety of biomedical applications in microscopy and beyond.
Ultra Low-Parameter Denoising: Trainable Bilateral Filter Layers in Computed Tomography
Jan 25, 2022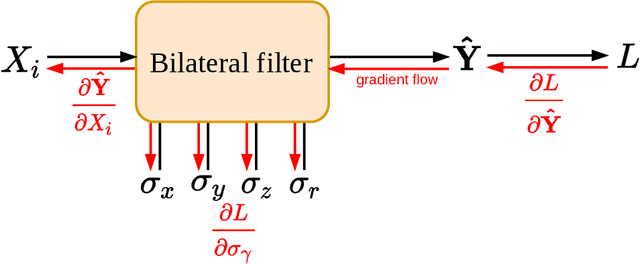
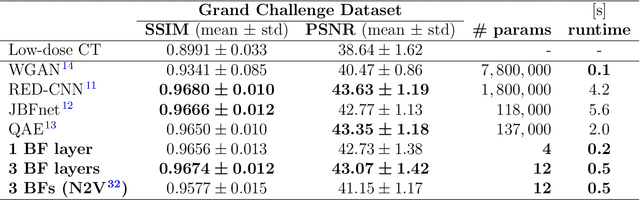
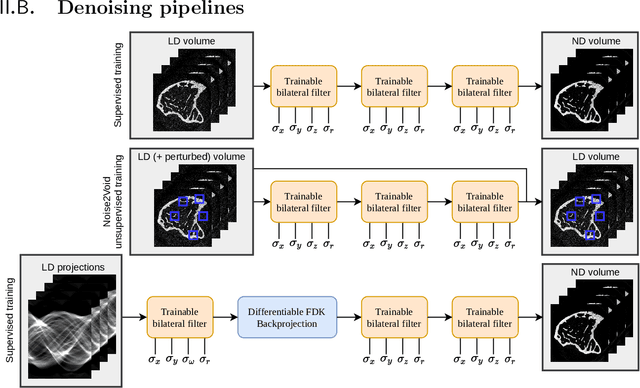
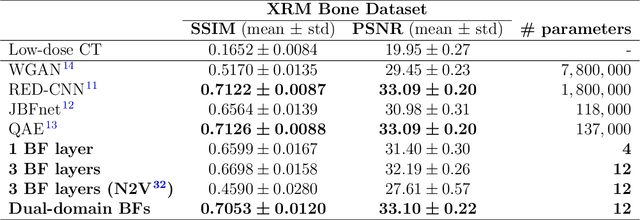
Abstract:Computed tomography is widely used as an imaging tool to visualize three-dimensional structures with expressive bone-soft tissue contrast. However, CT resolution and radiation dose are tightly entangled, highlighting the importance of low-dose CT combined with sophisticated denoising algorithms. Most data-driven denoising techniques are based on deep neural networks and, therefore, contain hundreds of thousands of trainable parameters, making them incomprehensible and prone to prediction failures. Developing understandable and robust denoising algorithms achieving state-of-the-art performance helps to minimize radiation dose while maintaining data integrity. This work presents an open-source CT denoising framework based on the idea of bilateral filtering. We propose a bilateral filter that can be incorporated into a deep learning pipeline and optimized in a purely data-driven way by calculating the gradient flow toward its hyperparameters and its input. Denoising in pure image-to-image pipelines and across different domains such as raw detector data and reconstructed volume, using a differentiable backprojection layer, is demonstrated. Although only using three spatial parameters and one range parameter per filter layer, the proposed denoising pipelines can compete with deep state-of-the-art denoising architectures with several hundred thousand parameters. Competitive denoising performance is achieved on x-ray microscope bone data (0.7053 and 33.10) and the 2016 Low Dose CT Grand Challenge dataset (0.9674 and 43.07) in terms of SSIM and PSNR. Due to the extremely low number of trainable parameters with well-defined effect, prediction reliance and data integrity is guaranteed at any time in the proposed pipelines, in contrast to most other deep learning-based denoising architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge