Sabrina Pechmann
Motion Compensation via Epipolar Consistency for In-Vivo X-Ray Microscopy
Mar 01, 2023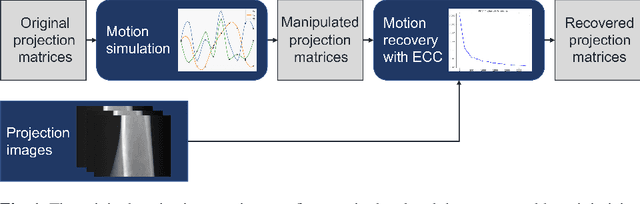
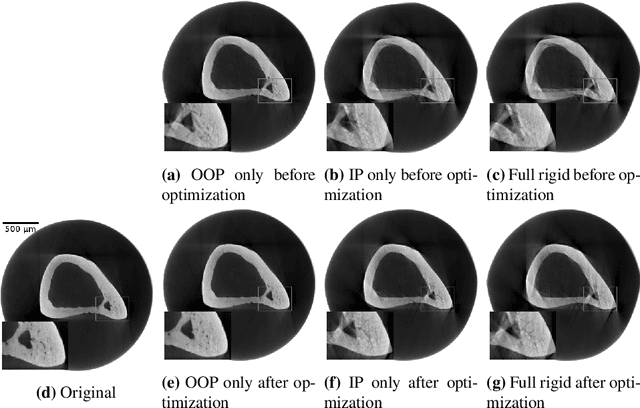
Abstract:Intravital X-ray microscopy (XRM) in preclinical mouse models is of vital importance for the identification of microscopic structural pathological changes in the bone which are characteristic of osteoporosis. The complexity of this method stems from the requirement for high-quality 3D reconstructions of the murine bones. However, respiratory motion and muscle relaxation lead to inconsistencies in the projection data which result in artifacts in uncompensated reconstructions. Motion compensation using epipolar consistency conditions (ECC) has previously shown good performance in clinical CT settings. Here, we explore whether such algorithms are suitable for correcting motion-corrupted XRM data. Different rigid motion patterns are simulated and the quality of the motion-compensated reconstructions is assessed. The method is able to restore microscopic features for out-of-plane motion, but artifacts remain for more realistic motion patterns including all six degrees of freedom of rigid motion. Therefore, ECC is valuable for the initial alignment of the projection data followed by further fine-tuning of motion parameters using a reconstruction-based method
Geometric Constraints Enable Self-Supervised Sinogram Inpainting in Sparse-View Tomography
Feb 13, 2023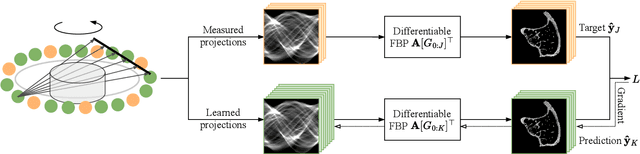
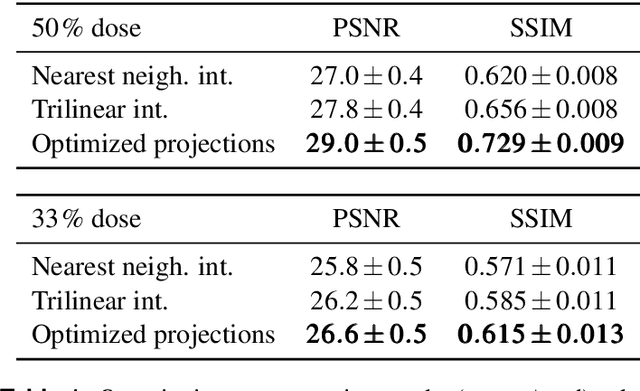
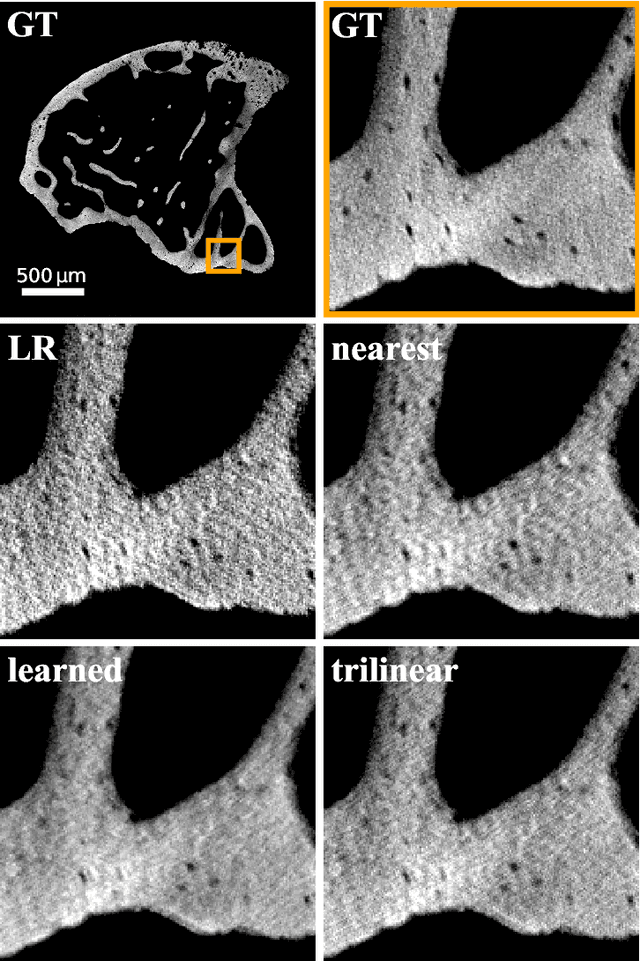
Abstract:The diagnostic quality of computed tomography (CT) scans is usually restricted by the induced patient dose, scan speed, and image quality. Sparse-angle tomographic scans reduce radiation exposure and accelerate data acquisition, but suffer from image artifacts and noise. Existing image processing algorithms can restore CT reconstruction quality but often require large training data sets or can not be used for truncated objects. This work presents a self-supervised projection inpainting method that allows learning missing projective views via gradient-based optimization. By reconstructing independent stacks of projection data, a self-supervised loss is calculated in the CT image domain and used to directly optimize projection image intensities to match the missing tomographic views constrained by the projection geometry. Our experiments on real X-ray microscope (XRM) tomographic mouse tibia bone scans show that our method improves reconstructions by 3.1-7.4%/7.7-17.6% in terms of PSNR/SSIM with respect to the interpolation baseline. Our approach is applicable as a flexible self-supervised projection inpainting tool for tomographic applications.
Noise2Contrast: Multi-Contrast Fusion Enables Self-Supervised Tomographic Image Denoising
Dec 09, 2022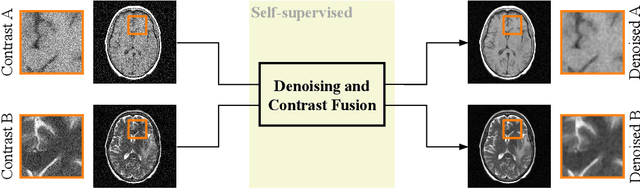
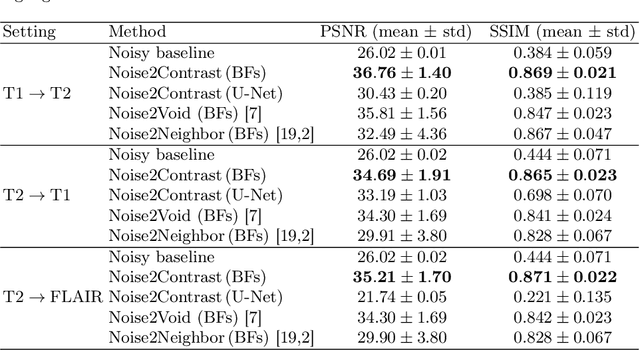
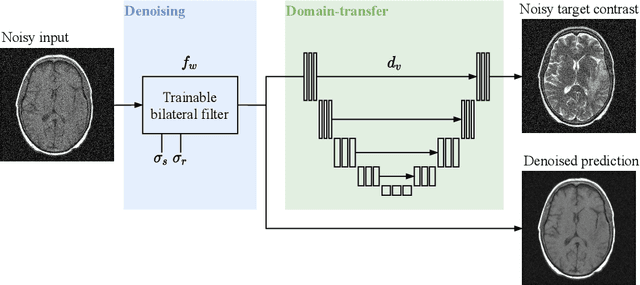
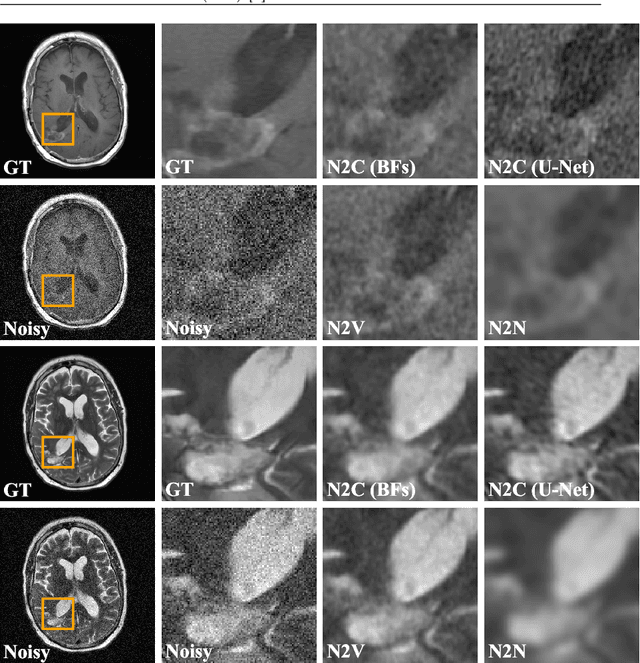
Abstract:Self-supervised image denoising techniques emerged as convenient methods that allow training denoising models without requiring ground-truth noise-free data. Existing methods usually optimize loss metrics that are calculated from multiple noisy realizations of similar images, e.g., from neighboring tomographic slices. However, those approaches fail to utilize the multiple contrasts that are routinely acquired in medical imaging modalities like MRI or dual-energy CT. In this work, we propose the new self-supervised training scheme Noise2Contrast that combines information from multiple measured image contrasts to train a denoising model. We stack denoising with domain-transfer operators to utilize the independent noise realizations of different image contrasts to derive a self-supervised loss. The trained denoising operator achieves convincing quantitative and qualitative results, outperforming state-of-the-art self-supervised methods by 4.7-11.0%/4.8-7.3% (PSNR/SSIM) on brain MRI data and by 43.6-50.5%/57.1-77.1% (PSNR/SSIM) on dual-energy CT X-ray microscopy data with respect to the noisy baseline. Our experiments on different real measured data sets indicate that Noise2Contrast training generalizes to other multi-contrast imaging modalities.
On the Benefit of Dual-domain Denoising in a Self-supervised Low-dose CT Setting
Nov 03, 2022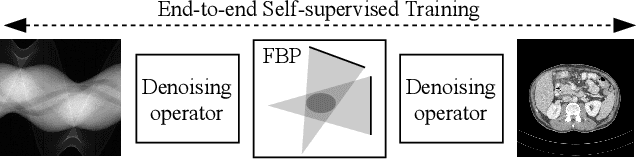
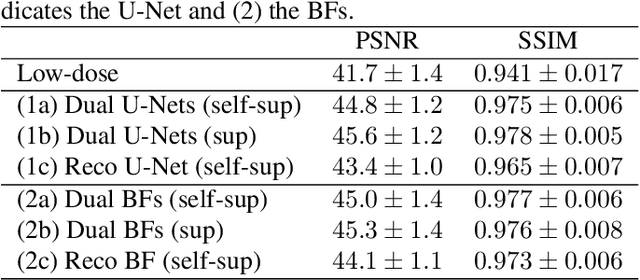
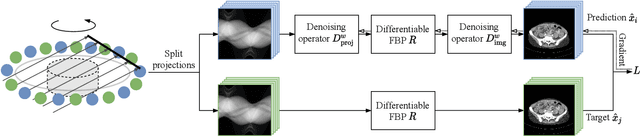
Abstract:Computed tomography (CT) is routinely used for three-dimensional non-invasive imaging. Numerous data-driven image denoising algorithms were proposed to restore image quality in low-dose acquisitions. However, considerably less research investigates methods already intervening in the raw detector data due to limited access to suitable projection data or correct reconstruction algorithms. In this work, we present an end-to-end trainable CT reconstruction pipeline that contains denoising operators in both the projection and the image domain and that are optimized simultaneously without requiring ground-truth high-dose CT data. Our experiments demonstrate that including an additional projection denoising operator improved the overall denoising performance by 82.4-94.1%/12.5-41.7% (PSNR/SSIM) on abdomen CT and 1.5-2.9%/0.4-0.5% (PSNR/SSIM) on XRM data relative to the low-dose baseline. We make our entire helical CT reconstruction framework publicly available that contains a raw projection rebinning step to render helical projection data suitable for differentiable fan-beam reconstruction operators and end-to-end learning.
Ultra Low-Parameter Denoising: Trainable Bilateral Filter Layers in Computed Tomography
Jan 25, 2022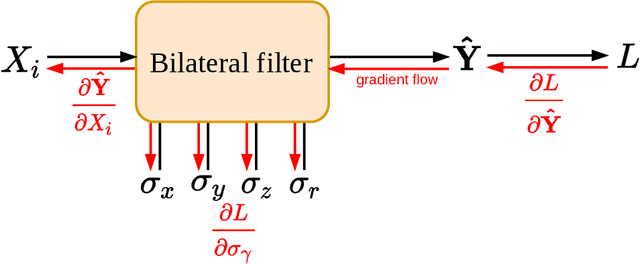
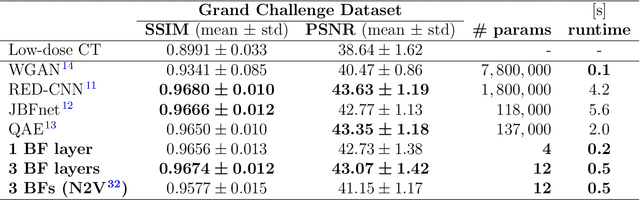
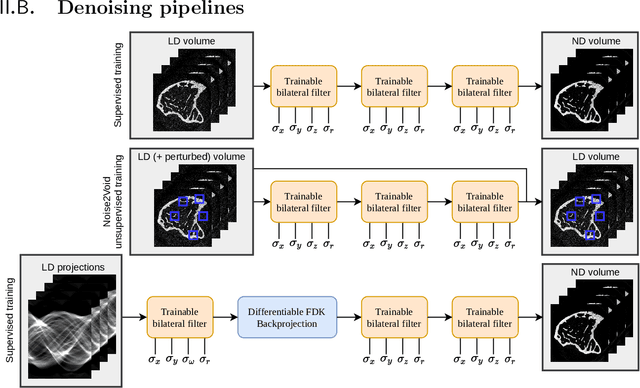
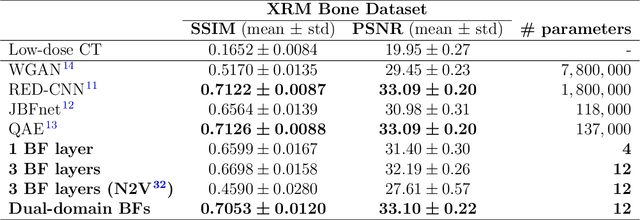
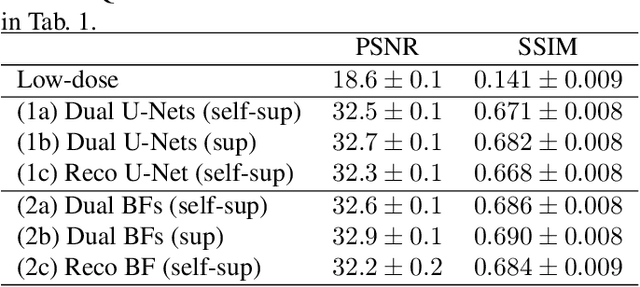
Abstract:Computed tomography is widely used as an imaging tool to visualize three-dimensional structures with expressive bone-soft tissue contrast. However, CT resolution and radiation dose are tightly entangled, highlighting the importance of low-dose CT combined with sophisticated denoising algorithms. Most data-driven denoising techniques are based on deep neural networks and, therefore, contain hundreds of thousands of trainable parameters, making them incomprehensible and prone to prediction failures. Developing understandable and robust denoising algorithms achieving state-of-the-art performance helps to minimize radiation dose while maintaining data integrity. This work presents an open-source CT denoising framework based on the idea of bilateral filtering. We propose a bilateral filter that can be incorporated into a deep learning pipeline and optimized in a purely data-driven way by calculating the gradient flow toward its hyperparameters and its input. Denoising in pure image-to-image pipelines and across different domains such as raw detector data and reconstructed volume, using a differentiable backprojection layer, is demonstrated. Although only using three spatial parameters and one range parameter per filter layer, the proposed denoising pipelines can compete with deep state-of-the-art denoising architectures with several hundred thousand parameters. Competitive denoising performance is achieved on x-ray microscope bone data (0.7053 and 33.10) and the 2016 Low Dose CT Grand Challenge dataset (0.9674 and 43.07) in terms of SSIM and PSNR. Due to the extremely low number of trainable parameters with well-defined effect, prediction reliance and data integrity is guaranteed at any time in the proposed pipelines, in contrast to most other deep learning-based denoising architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge