Yixing Huang
Oncology, University Hospital Erlangen, Friedrich-Alexander-University Erlangen-Nuremberg, 91054 Erlangen, Germany) These authors contributed equally: Zhisheng Wang, Haijun Yu Corresponding authors: Junning Cui, Fenglin Liu
A Self-supervised Multimodal Deep Learning Approach to Differentiate Post-radiotherapy Progression from Pseudoprogression in Glioblastoma
Feb 06, 2025



Abstract:Accurate differentiation of pseudoprogression (PsP) from True Progression (TP) following radiotherapy (RT) in glioblastoma (GBM) patients is crucial for optimal treatment planning. However, this task remains challenging due to the overlapping imaging characteristics of PsP and TP. This study therefore proposes a multimodal deep-learning approach utilizing complementary information from routine anatomical MR images, clinical parameters, and RT treatment planning information for improved predictive accuracy. The approach utilizes a self-supervised Vision Transformer (ViT) to encode multi-sequence MR brain volumes to effectively capture both global and local context from the high dimensional input. The encoder is trained in a self-supervised upstream task on unlabeled glioma MRI datasets from the open BraTS2021, UPenn-GBM, and UCSF-PDGM datasets to generate compact, clinically relevant representations from FLAIR and T1 post-contrast sequences. These encoded MR inputs are then integrated with clinical data and RT treatment planning information through guided cross-modal attention, improving progression classification accuracy. This work was developed using two datasets from different centers: the Burdenko Glioblastoma Progression Dataset (n = 59) for training and validation, and the GlioCMV progression dataset from the University Hospital Erlangen (UKER) (n = 20) for testing. The proposed method achieved an AUC of 75.3%, outperforming the current state-of-the-art data-driven approaches. Importantly, the proposed approach relies on readily available anatomical MRI sequences, clinical data, and RT treatment planning information, enhancing its clinical feasibility. The proposed approach addresses the challenge of limited data availability for PsP and TP differentiation and could allow for improved clinical decision-making and optimized treatment plans for GBM patients.
Exploring the Capabilities and Limitations of Large Language Models for Radiation Oncology Decision Support
Jan 04, 2025



Abstract:Thanks to the rapidly evolving integration of LLMs into decision-support tools, a significant transformation is happening across large-scale systems. Like other medical fields, the use of LLMs such as GPT-4 is gaining increasing interest in radiation oncology as well. An attempt to assess GPT-4's performance in radiation oncology was made via a dedicated 100-question examination on the highly specialized topic of radiation oncology physics, revealing GPT-4's superiority over other LLMs. GPT-4's performance on a broader field of clinical radiation oncology is further benchmarked by the ACR Radiation Oncology In-Training (TXIT) exam where GPT-4 achieved a high accuracy of 74.57%. Its performance on re-labelling structure names in accordance with the AAPM TG-263 report has also been benchmarked, achieving above 96% accuracies. Such studies shed light on the potential of LLMs in radiation oncology. As interest in the potential and constraints of LLMs in general healthcare applications continues to rise5, the capabilities and limitations of LLMs in radiation oncology decision support have not yet been fully explored.
* Officially published in the Red Journal
Task-Specific Data Preparation for Deep Learning to Reconstruct Structures of Interest from Severely Truncated CBCT Data
Sep 13, 2024



Abstract:Cone-beam computed tomography (CBCT) is widely used in interventional surgeries and radiation oncology. Due to the limited size of flat-panel detectors, anatomical structures might be missing outside the limited field-of-view (FOV), which restricts the clinical applications of CBCT systems. Recently, deep learning methods have been proposed to extend the FOV for multi-slice CT systems. However, in mobile CBCT system with a smaller FOV size, projection data is severely truncated and it is challenging for a network to restore all missing structures outside the FOV. In some applications, only certain structures outside the FOV are of interest, e.g., ribs in needle path planning for liver/lung cancer diagnosis. Therefore, a task-specific data preparation method is proposed in this work, which automatically let the network focus on structures of interest instead of all the structures. Our preliminary experiment shows that Pix2pixGAN with a conventional training has the risk to reconstruct false positive and false negative rib structures from severely truncated CBCT data, whereas Pix2pixGAN with the proposed task-specific training can reconstruct all the ribs reliably. The proposed method is promising to empower CBCT with more clinical applications.
Enhancing Cross-Modality Synthesis: Subvolume Merging for MRI-to-CT Conversion
Sep 09, 2024



Abstract:Providing more precise tissue attenuation information, synthetic computed tomography (sCT) generated from magnetic resonance imaging (MRI) contributes to improved radiation therapy treatment planning. In our study, we employ the advanced SwinUNETR framework for synthesizing CT from MRI images. Additionally, we introduce a three-dimensional subvolume merging technique in the prediction process. By selecting an optimal overlap percentage for adjacent subvolumes, stitching artifacts are effectively mitigated, leading to a decrease in the mean absolute error (MAE) between sCT and the labels from 52.65 HU to 47.75 HU. Furthermore, implementing a weight function with a gamma value of 0.9 results in the lowest MAE within the same overlap area. By setting the overlap percentage between 50% and 70%, we achieve a balance between image quality and computational efficiency.
Fine-Tuning a Local LLaMA-3 Large Language Model for Automated Privacy-Preserving Physician Letter Generation in Radiation Oncology
Aug 20, 2024



Abstract:Generating physician letters is a time-consuming task in daily clinical practice. This study investigates local fine-tuning of large language models (LLMs), specifically LLaMA models, for physician letter generation in a privacy-preserving manner within the field of radiation oncology. Our findings demonstrate that base LLaMA models, without fine-tuning, are inadequate for effectively generating physician letters. The QLoRA algorithm provides an efficient method for local intra-institutional fine-tuning of LLMs with limited computational resources (i.e., a single 48 GB GPU workstation within the hospital). The fine-tuned LLM successfully learns radiation oncology-specific information and generates physician letters in an institution-specific style. ROUGE scores of the generated summary reports highlight the superiority of the 8B LLaMA-3 model over the 13B LLaMA-2 model. Further multidimensional physician evaluations of 10 cases reveal that, although the fine-tuned LLaMA-3 model has limited capacity to generate content beyond the provided input data, it successfully generates salutations, diagnoses and treatment histories, recommendations for further treatment, and planned schedules. Overall, clinical benefit was rated highly by the clinical experts (average score of 3.44 on a 4-point scale). With careful physician review and correction, automated LLM-based physician letter generation has significant practical value.
Comprehensive Multimodal Deep Learning Survival Prediction Enabled by a Transformer Architecture: A Multicenter Study in Glioblastoma
May 21, 2024Abstract:Background: This research aims to improve glioblastoma survival prediction by integrating MR images, clinical and molecular-pathologic data in a transformer-based deep learning model, addressing data heterogeneity and performance generalizability. Method: We propose and evaluate a transformer-based non-linear and non-proportional survival prediction model. The model employs self-supervised learning techniques to effectively encode the high-dimensional MRI input for integration with non-imaging data using cross-attention. To demonstrate model generalizability, the model is assessed with the time-dependent concordance index (Cdt) in two training setups using three independent public test sets: UPenn-GBM, UCSF-PDGM, and RHUH-GBM, each comprising 378, 366, and 36 cases, respectively. Results: The proposed transformer model achieved promising performance for imaging as well as non-imaging data, effectively integrating both modalities for enhanced performance (UPenn-GBM test-set, imaging Cdt 0.645, multimodal Cdt 0.707) while outperforming state-of-the-art late-fusion 3D-CNN-based models. Consistent performance was observed across the three independent multicenter test sets with Cdt values of 0.707 (UPenn-GBM, internal test set), 0.672 (UCSF-PDGM, first external test set) and 0.618 (RHUH-GBM, second external test set). The model achieved significant discrimination between patients with favorable and unfavorable survival for all three datasets (logrank p 1.9\times{10}^{-8}, 9.7\times{10}^{-3}, and 1.2\times{10}^{-2}). Conclusions: The proposed transformer-based survival prediction model integrates complementary information from diverse input modalities, contributing to improved glioblastoma survival prediction compared to state-of-the-art methods. Consistent performance was observed across institutions supporting model generalizability.
Multicenter Privacy-Preserving Model Training for Deep Learning Brain Metastases Autosegmentation
May 17, 2024



Abstract:Objectives: This work aims to explore the impact of multicenter data heterogeneity on deep learning brain metastases (BM) autosegmentation performance, and assess the efficacy of an incremental transfer learning technique, namely learning without forgetting (LWF), to improve model generalizability without sharing raw data. Materials and methods: A total of six BM datasets from University Hospital Erlangen (UKER), University Hospital Zurich (USZ), Stanford, UCSF, NYU and BraTS Challenge 2023 on BM segmentation were used for this evaluation. First, the multicenter performance of a convolutional neural network (DeepMedic) for BM autosegmentation was established for exclusive single-center training and for training on pooled data, respectively. Subsequently bilateral collaboration was evaluated, where a UKER pretrained model is shared to another center for further training using transfer learning (TL) either with or without LWF. Results: For single-center training, average F1 scores of BM detection range from 0.625 (NYU) to 0.876 (UKER) on respective single-center test data. Mixed multicenter training notably improves F1 scores at Stanford and NYU, with negligible improvement at other centers. When the UKER pretrained model is applied to USZ, LWF achieves a higher average F1 score (0.839) than naive TL (0.570) and single-center training (0.688) on combined UKER and USZ test data. Naive TL improves sensitivity and contouring accuracy, but compromises precision. Conversely, LWF demonstrates commendable sensitivity, precision and contouring accuracy. When applied to Stanford, similar performance was observed. Conclusion: Data heterogeneity results in varying performance in BM autosegmentation, posing challenges to model generalizability. LWF is a promising approach to peer-to-peer privacy-preserving model training.
Reference-Free Multi-Modality Volume Registration of X-Ray Microscopy and Light-Sheet Fluorescence Microscopy
Apr 23, 2024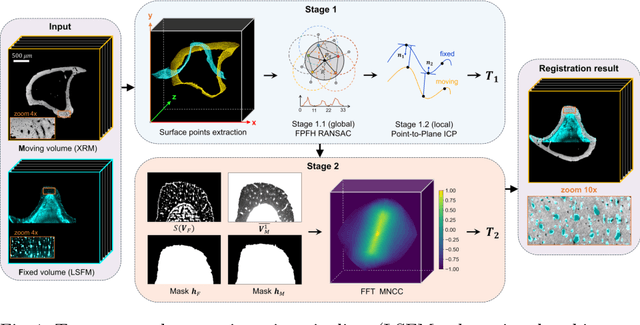
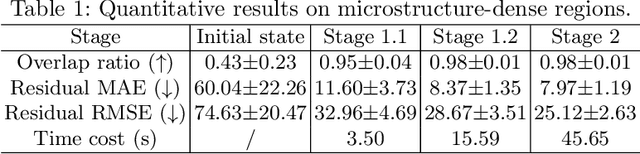
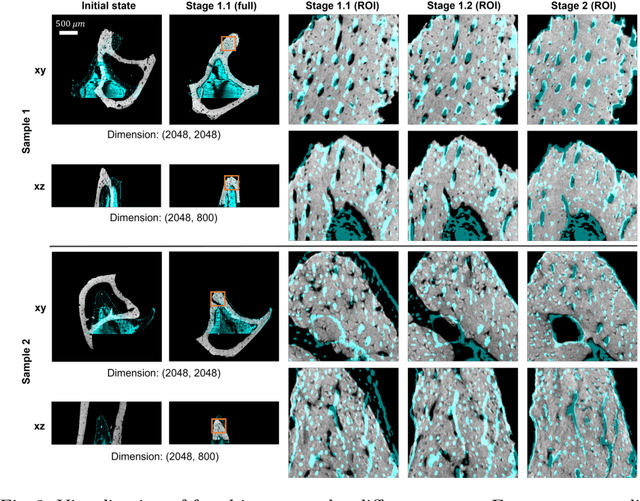
Abstract:Recently, X-ray microscopy (XRM) and light-sheet fluorescence microscopy (LSFM) have emerged as two pivotal imaging tools in preclinical research on bone remodeling diseases, offering micrometer-level resolution. Integrating these complementary modalities provides a holistic view of bone microstructures, facilitating function-oriented volume analysis across different disease cycles. However, registering such independently acquired large-scale volumes is extremely challenging under real and reference-free scenarios. This paper presents a fast two-stage pipeline for volume registration of XRM and LSFM. The first stage extracts the surface features and employs two successive point cloud-based methods for coarse alignment. The second stage fine-tunes the initial alignment using a modified cross-correlation method, ensuring precise volumetric registration. Moreover, we propose residual similarity as a novel metric to assess the alignment of two complementary modalities. The results imply robust gradual improvement across the stages. In the end, all correlating microstructures, particularly lacunae in XRM and bone cells in LSFM, are precisely matched, enabling new insights into bone diseases like osteoporosis which are a substantial burden in aging societies.
EAGLE: An Edge-Aware Gradient Localization Enhanced Loss for CT Image Reconstruction
Mar 15, 2024Abstract:Computed Tomography (CT) image reconstruction is crucial for accurate diagnosis and deep learning approaches have demonstrated significant potential in improving reconstruction quality. However, the choice of loss function profoundly affects the reconstructed images. Traditional mean squared error loss often produces blurry images lacking fine details, while alternatives designed to improve may introduce structural artifacts or other undesirable effects. To address these limitations, we propose Eagle-Loss, a novel loss function designed to enhance the visual quality of CT image reconstructions. Eagle-Loss applies spectral analysis of localized features within gradient changes to enhance sharpness and well-defined edges. We evaluated Eagle-Loss on two public datasets across low-dose CT reconstruction and CT field-of-view extension tasks. Our results show that Eagle-Loss consistently improves the visual quality of reconstructed images, surpassing state-of-the-art methods across various network architectures. Code and data are available at \url{https://github.com/sypsyp97/Eagle_Loss}.
Two-View Topogram-Based Anatomy-Guided CT Reconstruction for Prospective Risk Minimization
Jan 23, 2024Abstract:To facilitate a prospective estimation of CT effective dose and risk minimization process, a prospective spatial dose estimation and the known anatomical structures are expected. To this end, a CT reconstruction method is required to reconstruct CT volumes from as few projections as possible, i.e. by using the topograms, with anatomical structures as correct as possible. In this work, an optimized CT reconstruction model based on a generative adversarial network (GAN) is proposed. The GAN is trained to reconstruct 3D volumes from an anterior-posterior and a lateral CT projection. To enhance anatomical structures, a pre-trained organ segmentation network and the 3D perceptual loss are applied during the training phase, so that the model can then generate both organ-enhanced CT volume and the organ segmentation mask. The proposed method can reconstruct CT volumes with PSNR of 26.49, RMSE of 196.17, and SSIM of 0.64, compared to 26.21, 201.55 and 0.63 using the baseline method. In terms of the anatomical structure, the proposed method effectively enhances the organ shape and boundary and allows for a straight-forward identification of the relevant anatomical structures. We note that conventional reconstruction metrics fail to indicate the enhancement of anatomical structures. In addition to such metrics, the evaluation is expanded with assessing the organ segmentation performance. The average organ dice of the proposed method is 0.71 compared with 0.63 in baseline model, indicating the enhancement of anatomical structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge