Philipp Schubert
Benchmarking GPT-5 in Radiation Oncology: Measurable Gains, but Persistent Need for Expert Oversight
Aug 29, 2025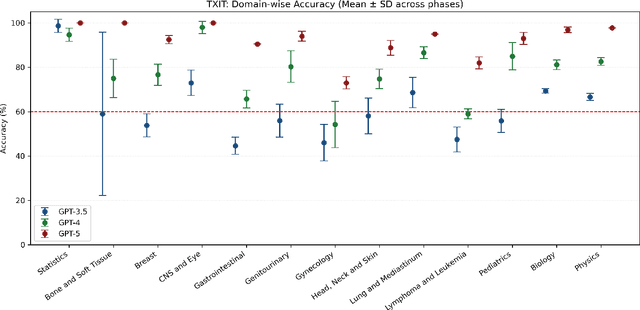
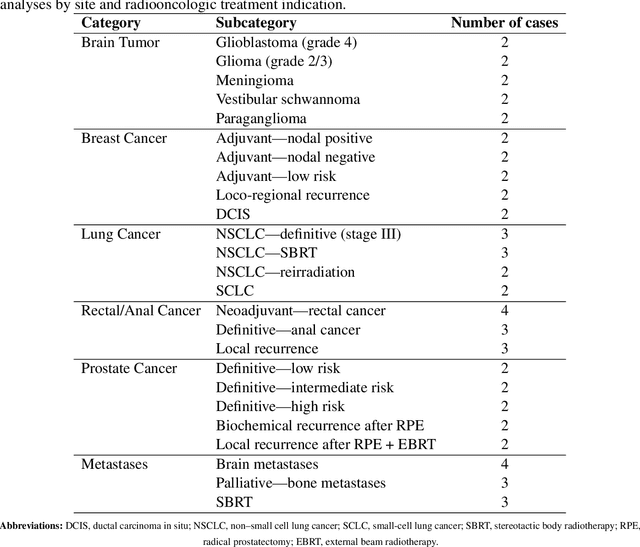
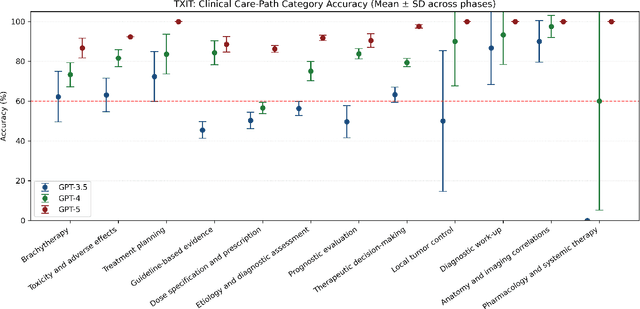
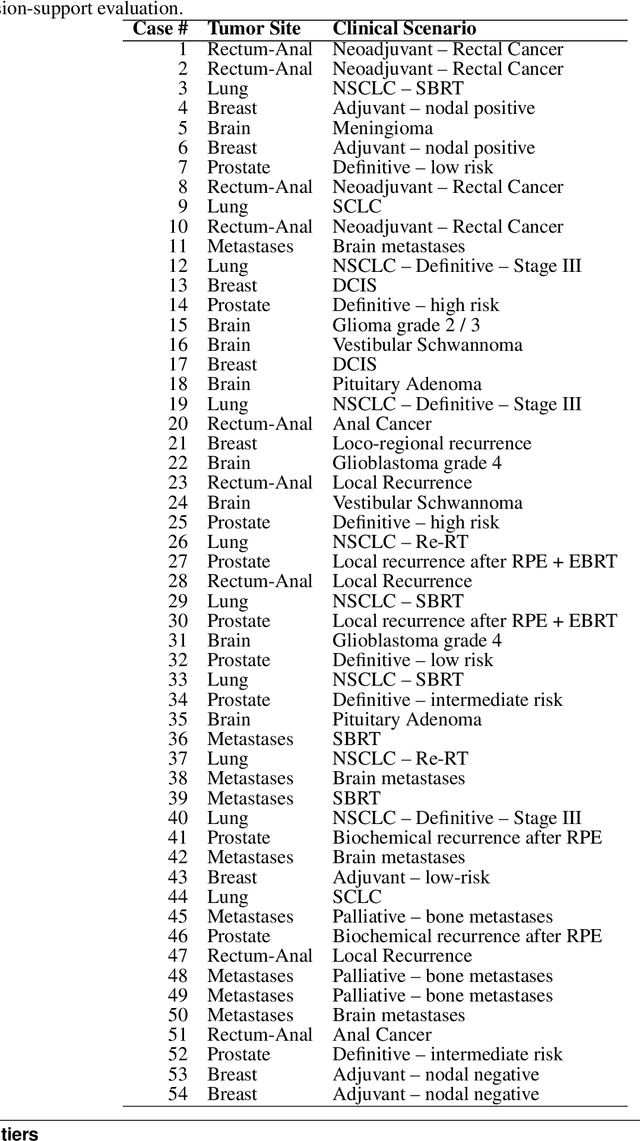
Abstract:Introduction: Large language models (LLM) have shown great potential in clinical decision support. GPT-5 is a novel LLM system that has been specifically marketed towards oncology use. Methods: Performance was assessed using two complementary benchmarks: (i) the ACR Radiation Oncology In-Training Examination (TXIT, 2021), comprising 300 multiple-choice items, and (ii) a curated set of 60 authentic radiation oncologic vignettes representing diverse disease sites and treatment indications. For the vignette evaluation, GPT-5 was instructed to generate concise therapeutic plans. Four board-certified radiation oncologists rated correctness, comprehensiveness, and hallucinations. Inter-rater reliability was quantified using Fleiss' \k{appa}. Results: On the TXIT benchmark, GPT-5 achieved a mean accuracy of 92.8%, outperforming GPT-4 (78.8%) and GPT-3.5 (62.1%). Domain-specific gains were most pronounced in Dose and Diagnosis. In the vignette evaluation, GPT-5's treatment recommendations were rated highly for correctness (mean 3.24/4, 95% CI: 3.11-3.38) and comprehensiveness (3.59/4, 95% CI: 3.49-3.69). Hallucinations were rare with no case reaching majority consensus for their presence. Inter-rater agreement was low (Fleiss' \k{appa} 0.083 for correctness), reflecting inherent variability in clinical judgment. Errors clustered in complex scenarios requiring precise trial knowledge or detailed clinical adaptation. Discussion: GPT-5 clearly outperformed prior model variants on the radiation oncology multiple-choice benchmark. Although GPT-5 exhibited favorable performance in generating real-world radiation oncology treatment recommendations, correctness ratings indicate room for further improvement. While hallucinations were infrequent, the presence of substantive errors underscores that GPT-5-generated recommendations require rigorous expert oversight before clinical implementation.
Fine-Tuning a Local LLaMA-3 Large Language Model for Automated Privacy-Preserving Physician Letter Generation in Radiation Oncology
Aug 20, 2024



Abstract:Generating physician letters is a time-consuming task in daily clinical practice. This study investigates local fine-tuning of large language models (LLMs), specifically LLaMA models, for physician letter generation in a privacy-preserving manner within the field of radiation oncology. Our findings demonstrate that base LLaMA models, without fine-tuning, are inadequate for effectively generating physician letters. The QLoRA algorithm provides an efficient method for local intra-institutional fine-tuning of LLMs with limited computational resources (i.e., a single 48 GB GPU workstation within the hospital). The fine-tuned LLM successfully learns radiation oncology-specific information and generates physician letters in an institution-specific style. ROUGE scores of the generated summary reports highlight the superiority of the 8B LLaMA-3 model over the 13B LLaMA-2 model. Further multidimensional physician evaluations of 10 cases reveal that, although the fine-tuned LLaMA-3 model has limited capacity to generate content beyond the provided input data, it successfully generates salutations, diagnoses and treatment histories, recommendations for further treatment, and planned schedules. Overall, clinical benefit was rated highly by the clinical experts (average score of 3.44 on a 4-point scale). With careful physician review and correction, automated LLM-based physician letter generation has significant practical value.
The Segment Anything foundation model achieves favorable brain tumor autosegmentation accuracy on MRI to support radiotherapy treatment planning
Apr 16, 2023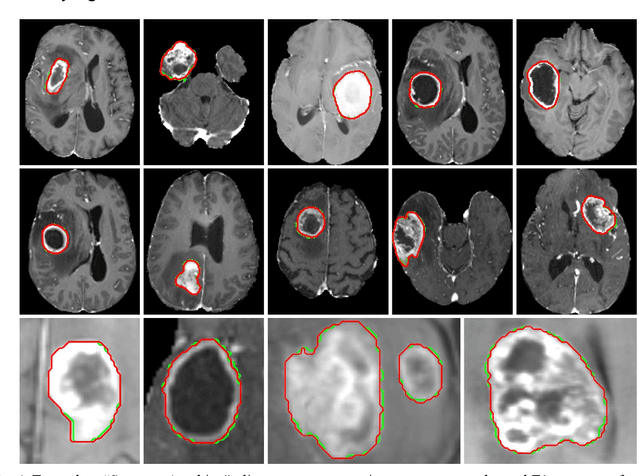
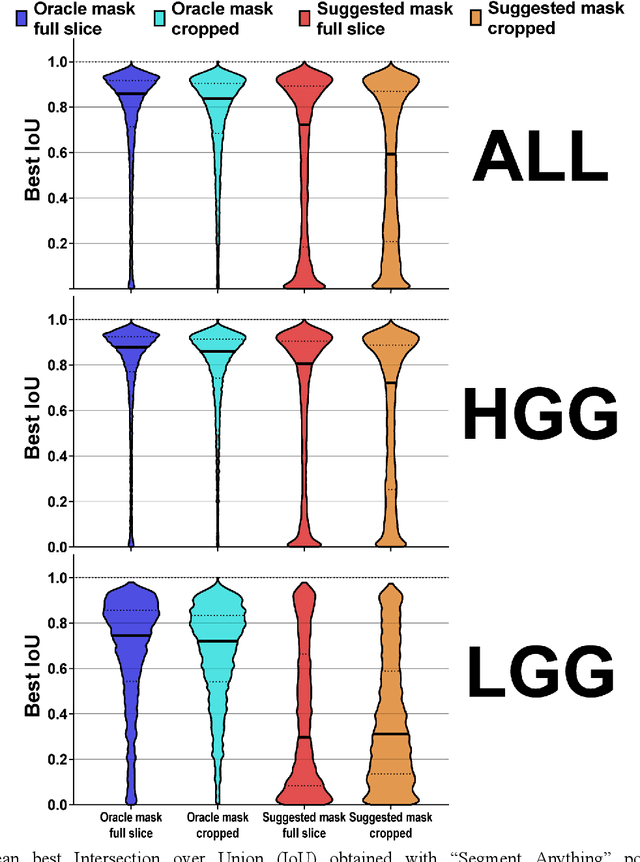
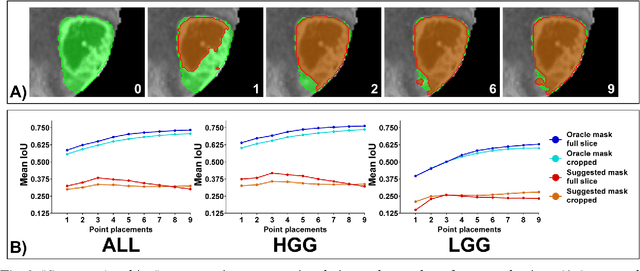
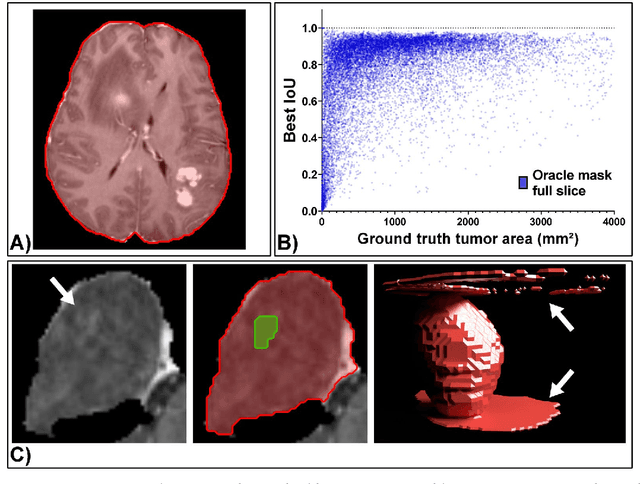
Abstract:Background: Tumor segmentation in MRI is crucial in radiotherapy (RT) treatment planning for brain tumor patients. Segment anything (SA), a novel promptable foundation model for autosegmentation, has shown high accuracy for multiple segmentation tasks but was not evaluated on medical datasets yet. Methods: SA was evaluated in a point-to-mask task for glioma brain tumor autosegmentation on 16744 transversal slices from 369 MRI datasets (BraTS 2020). Up to 9 point prompts were placed per slice. Tumor core (enhancing tumor + necrotic core) was segmented on contrast-enhanced T1w sequences. Out of the 3 masks predicted by SA, accuracy was evaluated for the mask with the highest calculated IoU (oracle mask) and with highest model predicted IoU (suggested mask). In addition to assessing SA on whole MRI slices, SA was also evaluated on images cropped to the tumor (max. 3D extent + 2 cm). Results: Mean best IoU (mbIoU) using oracle mask on full MRI slices was 0.762 (IQR 0.713-0.917). Best 2D mask was achieved after a mean of 6.6 point prompts (IQR 5-9). Segmentation accuracy was significantly better for high- compared to low-grade glioma cases (mbIoU 0.789 vs. 0.668). Accuracy was worse using MRI slices cropped to the tumor (mbIoU 0.759) and was much worse using suggested mask (full slices 0.572). For all experiments, accuracy was low on peripheral slices with few tumor voxels (mbIoU, <300: 0.537 vs. >=300: 0.841). Stacking best oracle segmentations from full axial MRI slices, mean 3D DSC for tumor core was 0.872, which was improved to 0.919 by combining axial, sagittal and coronal masks. Conclusions: The Segment Anything foundation model, while trained on photos, can achieve high zero-shot accuracy for glioma brain tumor segmentation on MRI slices. The results suggest that Segment Anything can accelerate and facilitate RT treatment planning, when properly integrated in a clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge