Thomas Weissmann
Benchmarking GPT-5 in Radiation Oncology: Measurable Gains, but Persistent Need for Expert Oversight
Aug 29, 2025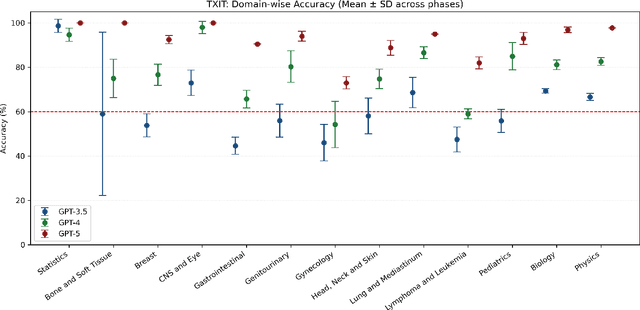
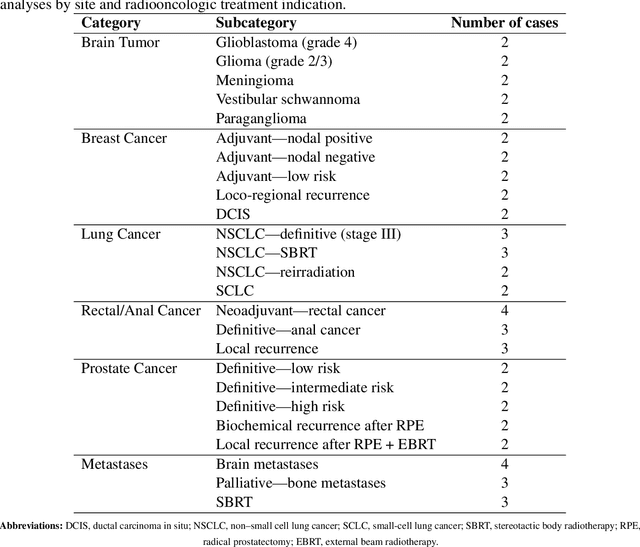
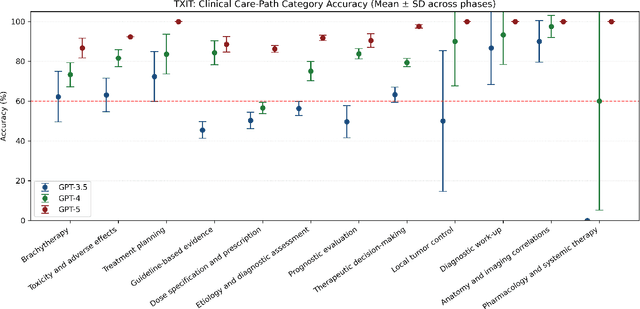
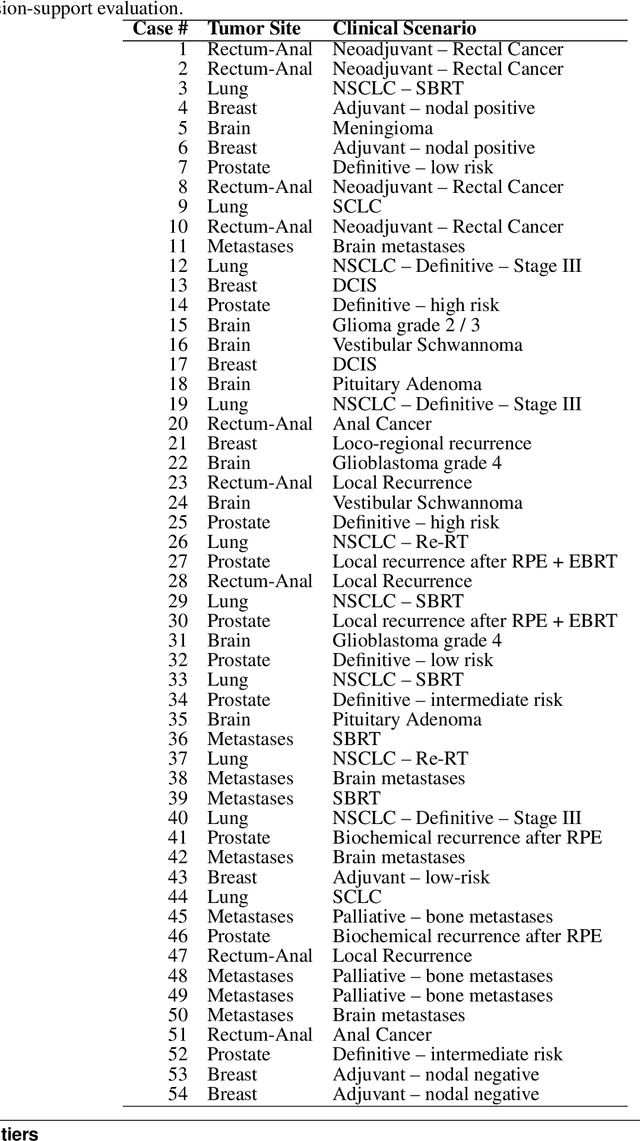
Abstract:Introduction: Large language models (LLM) have shown great potential in clinical decision support. GPT-5 is a novel LLM system that has been specifically marketed towards oncology use. Methods: Performance was assessed using two complementary benchmarks: (i) the ACR Radiation Oncology In-Training Examination (TXIT, 2021), comprising 300 multiple-choice items, and (ii) a curated set of 60 authentic radiation oncologic vignettes representing diverse disease sites and treatment indications. For the vignette evaluation, GPT-5 was instructed to generate concise therapeutic plans. Four board-certified radiation oncologists rated correctness, comprehensiveness, and hallucinations. Inter-rater reliability was quantified using Fleiss' \k{appa}. Results: On the TXIT benchmark, GPT-5 achieved a mean accuracy of 92.8%, outperforming GPT-4 (78.8%) and GPT-3.5 (62.1%). Domain-specific gains were most pronounced in Dose and Diagnosis. In the vignette evaluation, GPT-5's treatment recommendations were rated highly for correctness (mean 3.24/4, 95% CI: 3.11-3.38) and comprehensiveness (3.59/4, 95% CI: 3.49-3.69). Hallucinations were rare with no case reaching majority consensus for their presence. Inter-rater agreement was low (Fleiss' \k{appa} 0.083 for correctness), reflecting inherent variability in clinical judgment. Errors clustered in complex scenarios requiring precise trial knowledge or detailed clinical adaptation. Discussion: GPT-5 clearly outperformed prior model variants on the radiation oncology multiple-choice benchmark. Although GPT-5 exhibited favorable performance in generating real-world radiation oncology treatment recommendations, correctness ratings indicate room for further improvement. While hallucinations were infrequent, the presence of substantive errors underscores that GPT-5-generated recommendations require rigorous expert oversight before clinical implementation.
Fine-Tuning a Local LLaMA-3 Large Language Model for Automated Privacy-Preserving Physician Letter Generation in Radiation Oncology
Aug 20, 2024



Abstract:Generating physician letters is a time-consuming task in daily clinical practice. This study investigates local fine-tuning of large language models (LLMs), specifically LLaMA models, for physician letter generation in a privacy-preserving manner within the field of radiation oncology. Our findings demonstrate that base LLaMA models, without fine-tuning, are inadequate for effectively generating physician letters. The QLoRA algorithm provides an efficient method for local intra-institutional fine-tuning of LLMs with limited computational resources (i.e., a single 48 GB GPU workstation within the hospital). The fine-tuned LLM successfully learns radiation oncology-specific information and generates physician letters in an institution-specific style. ROUGE scores of the generated summary reports highlight the superiority of the 8B LLaMA-3 model over the 13B LLaMA-2 model. Further multidimensional physician evaluations of 10 cases reveal that, although the fine-tuned LLaMA-3 model has limited capacity to generate content beyond the provided input data, it successfully generates salutations, diagnoses and treatment histories, recommendations for further treatment, and planned schedules. Overall, clinical benefit was rated highly by the clinical experts (average score of 3.44 on a 4-point scale). With careful physician review and correction, automated LLM-based physician letter generation has significant practical value.
Comprehensive Multimodal Deep Learning Survival Prediction Enabled by a Transformer Architecture: A Multicenter Study in Glioblastoma
May 21, 2024Abstract:Background: This research aims to improve glioblastoma survival prediction by integrating MR images, clinical and molecular-pathologic data in a transformer-based deep learning model, addressing data heterogeneity and performance generalizability. Method: We propose and evaluate a transformer-based non-linear and non-proportional survival prediction model. The model employs self-supervised learning techniques to effectively encode the high-dimensional MRI input for integration with non-imaging data using cross-attention. To demonstrate model generalizability, the model is assessed with the time-dependent concordance index (Cdt) in two training setups using three independent public test sets: UPenn-GBM, UCSF-PDGM, and RHUH-GBM, each comprising 378, 366, and 36 cases, respectively. Results: The proposed transformer model achieved promising performance for imaging as well as non-imaging data, effectively integrating both modalities for enhanced performance (UPenn-GBM test-set, imaging Cdt 0.645, multimodal Cdt 0.707) while outperforming state-of-the-art late-fusion 3D-CNN-based models. Consistent performance was observed across the three independent multicenter test sets with Cdt values of 0.707 (UPenn-GBM, internal test set), 0.672 (UCSF-PDGM, first external test set) and 0.618 (RHUH-GBM, second external test set). The model achieved significant discrimination between patients with favorable and unfavorable survival for all three datasets (logrank p 1.9\times{10}^{-8}, 9.7\times{10}^{-3}, and 1.2\times{10}^{-2}). Conclusions: The proposed transformer-based survival prediction model integrates complementary information from diverse input modalities, contributing to improved glioblastoma survival prediction compared to state-of-the-art methods. Consistent performance was observed across institutions supporting model generalizability.
Benchmarking ChatGPT-4 on ACR Radiation Oncology In-Training Exam (TXIT): Potentials and Challenges for AI-Assisted Medical Education and Decision Making in Radiation Oncology
Apr 24, 2023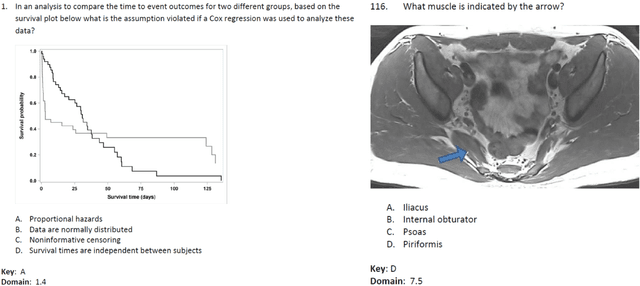
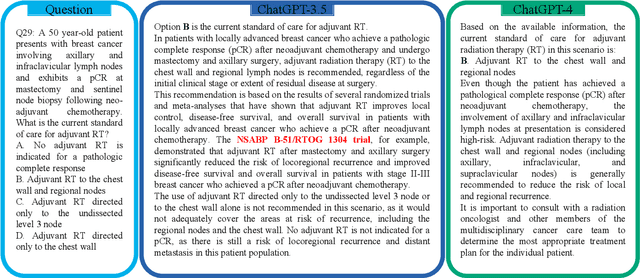
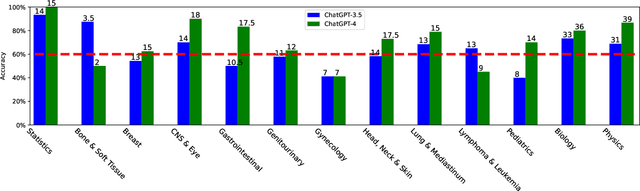
Abstract:The potential of large language models in medicine for education and decision making purposes has been demonstrated as they achieve decent scores on medical exams such as the United States Medical Licensing Exam (USMLE) and the MedQA exam. In this work, we evaluate the performance of ChatGPT-3.5 and ChatGPT-4 in the specialized field of radiation oncology using the 38th American College of Radiology (ACR) radiation oncology in-training exam (TXIT). ChatGPT-3.5 and ChatGPT-4 have achieved the scores of 63.65% and 74.57%, respectively, highlighting the advantage of the latest ChatGPT-4 model. Based on the TXIT exam, ChatGPT-4's strong and weak areas in radiation oncology are identified to some extent. Specifically, ChatGPT-4 demonstrates good knowledge of statistics, CNS & eye, pediatrics, biology, and physics but has limitations in bone & soft tissue and gynecology, as per the ACR knowledge domain. Regarding clinical care paths, ChatGPT-4 performs well in diagnosis, prognosis, and toxicity but lacks proficiency in topics related to brachytherapy and dosimetry, as well as in-depth questions from clinical trials. While ChatGPT-4 is not yet suitable for clinical decision making in radiation oncology, it has the potential to assist in medical education for the general public and cancer patients. With further fine-tuning, it could assist radiation oncologists in recommending treatment decisions for challenging clinical cases based on the latest guidelines and the existing gray zone database.
The Segment Anything foundation model achieves favorable brain tumor autosegmentation accuracy on MRI to support radiotherapy treatment planning
Apr 16, 2023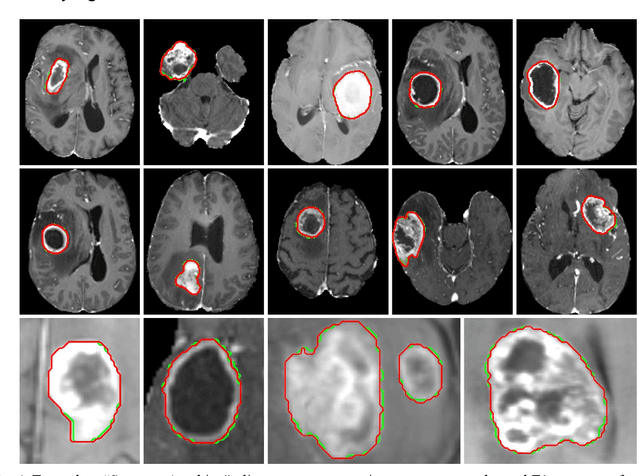
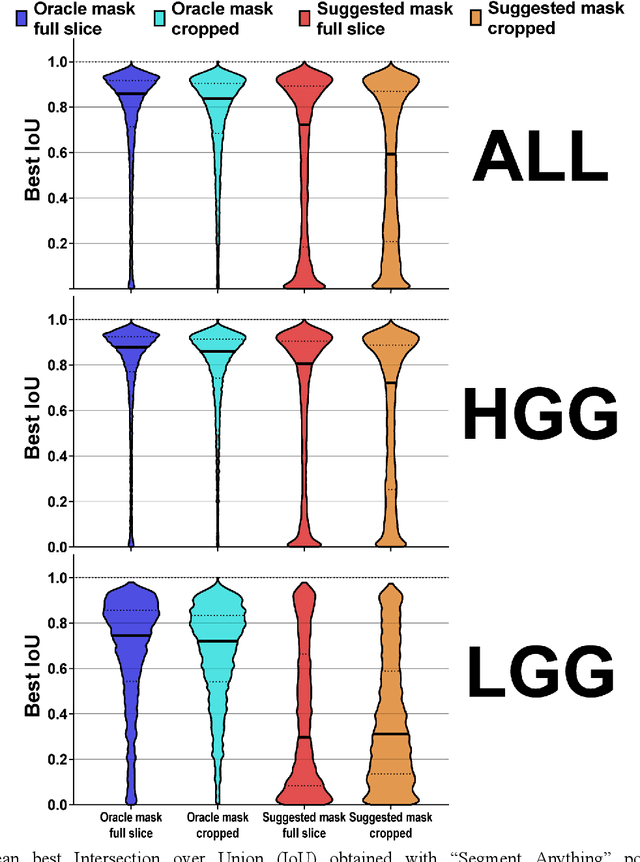
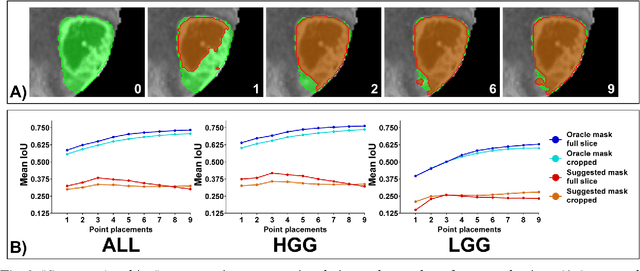
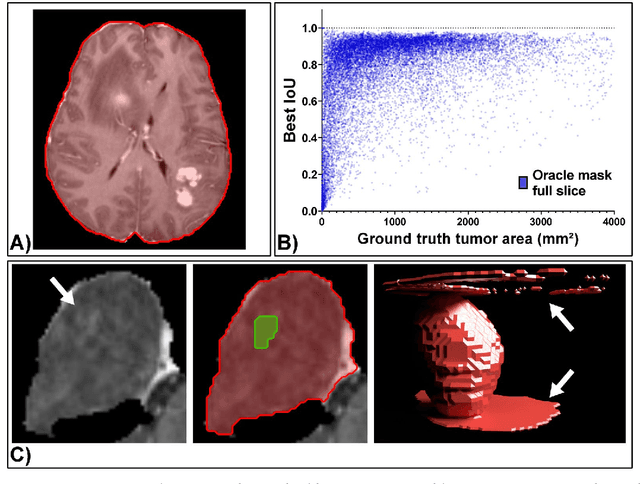
Abstract:Background: Tumor segmentation in MRI is crucial in radiotherapy (RT) treatment planning for brain tumor patients. Segment anything (SA), a novel promptable foundation model for autosegmentation, has shown high accuracy for multiple segmentation tasks but was not evaluated on medical datasets yet. Methods: SA was evaluated in a point-to-mask task for glioma brain tumor autosegmentation on 16744 transversal slices from 369 MRI datasets (BraTS 2020). Up to 9 point prompts were placed per slice. Tumor core (enhancing tumor + necrotic core) was segmented on contrast-enhanced T1w sequences. Out of the 3 masks predicted by SA, accuracy was evaluated for the mask with the highest calculated IoU (oracle mask) and with highest model predicted IoU (suggested mask). In addition to assessing SA on whole MRI slices, SA was also evaluated on images cropped to the tumor (max. 3D extent + 2 cm). Results: Mean best IoU (mbIoU) using oracle mask on full MRI slices was 0.762 (IQR 0.713-0.917). Best 2D mask was achieved after a mean of 6.6 point prompts (IQR 5-9). Segmentation accuracy was significantly better for high- compared to low-grade glioma cases (mbIoU 0.789 vs. 0.668). Accuracy was worse using MRI slices cropped to the tumor (mbIoU 0.759) and was much worse using suggested mask (full slices 0.572). For all experiments, accuracy was low on peripheral slices with few tumor voxels (mbIoU, <300: 0.537 vs. >=300: 0.841). Stacking best oracle segmentations from full axial MRI slices, mean 3D DSC for tumor core was 0.872, which was improved to 0.919 by combining axial, sagittal and coronal masks. Conclusions: The Segment Anything foundation model, while trained on photos, can achieve high zero-shot accuracy for glioma brain tumor segmentation on MRI slices. The results suggest that Segment Anything can accelerate and facilitate RT treatment planning, when properly integrated in a clinical application.
Deep Learning for automatic head and neck lymph node level delineation
Aug 28, 2022
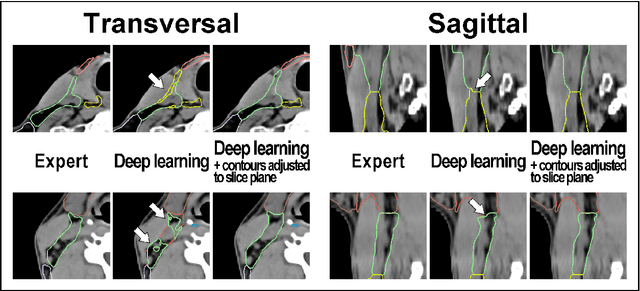
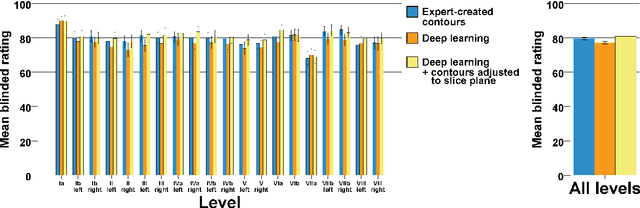
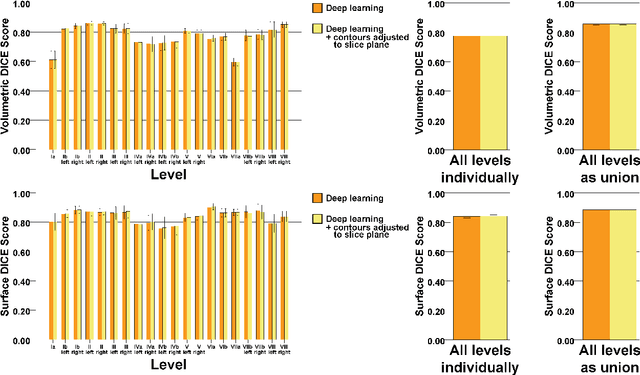
Abstract:Background: Deep learning-based head and neck lymph node level (HN_LNL) autodelineation is of high relevance to radiotherapy research and clinical treatment planning but still understudied in academic literature. Methods: An expert-delineated cohort of 35 planning CTs was used for training of an nnU-net 3D-fullres/2D-ensemble model for autosegmentation of 20 different HN_LNL. Validation was performed in an independent test set (n=20). In a completely blinded evaluation, 3 clinical experts rated the quality of deep learning autosegmentations in a head-to-head comparison with expert-created contours. For a subgroup of 10 cases, intraobserver variability was compared to deep learning autosegmentation performance. The effect of autocontour consistency with CT slice plane orientation on geometric accuracy and expert rating was investigated. Results: Mean blinded expert rating per level was significantly better for deep learning segmentations with CT slice plane adjustment than for expert-created contours (81.0 vs. 79.6, p<0.001), but deep learning segmentations without slice plane adjustment were rated significantly worse than expert-created contours (77.2 vs. 79.6, p<0.001). Geometric accuracy of deep learning segmentations was non-different from intraobserver variability (mean Dice per level, 0.78 vs. 0.77, p=0.064) with variance in accuracy between levels being improved (p<0.001). Clinical significance of contour consistency with CT slice plane orientation was not represented by geometric accuracy metrics (Dice, 0.78 vs. 0.78, p=0.572) Conclusions: We show that a nnU-net 3D-fullres/2D-ensemble model can be used for highly accurate autodelineation of HN_LNL using only a limited training dataset that is ideally suited for large-scale standardized autodelineation of HN_LNL in the research setting. Geometric accuracy metrics are only an imperfect surrogate for blinded expert rating.
Deep learning for brain metastasis detection and segmentation in longitudinal MRI data
Dec 28, 2021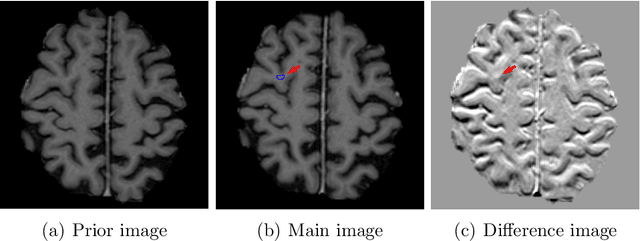
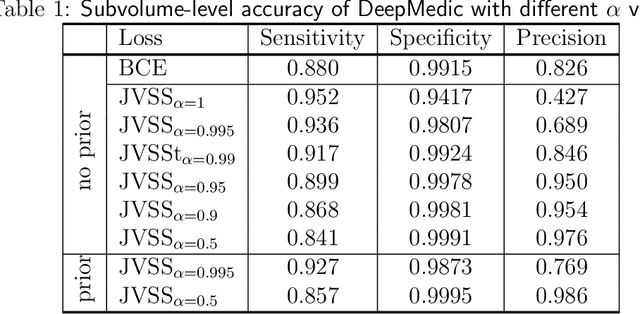
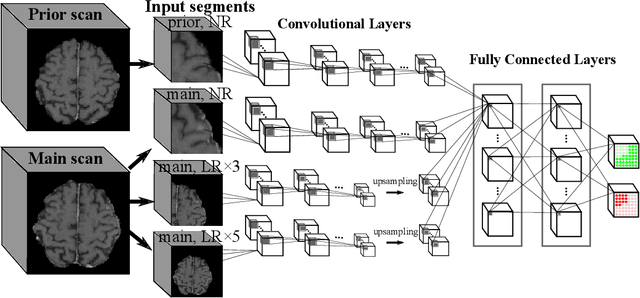
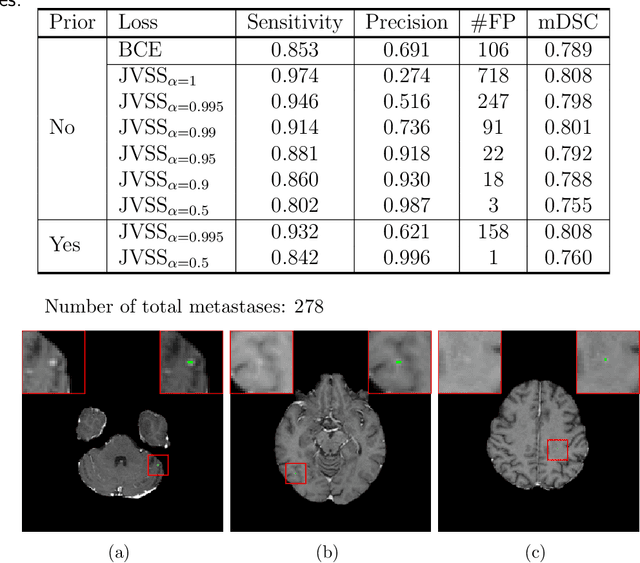
Abstract:Brain metastases occur frequently in patients with metastatic cancer. Early and accurate detection of brain metastases is very essential for treatment planning and prognosis in radiation therapy. To improve brain metastasis detection performance with deep learning, a custom detection loss called volume-level sensitivity-specificity (VSS) is proposed, which rates individual metastasis detection sensitivity and specificity in (sub-)volume levels. As sensitivity and precision are always a trade-off in a metastasis level, either a high sensitivity or a high precision can be achieved by adjusting the weights in the VSS loss without decline in dice score coefficient for segmented metastases. To reduce metastasis-like structures being detected as false positive metastases, a temporal prior volume is proposed as an additional input of the neural network. Our proposed VSS loss improves the sensitivity of brain metastasis detection, increasing the sensitivity from 86.7% to 95.5%. Alternatively, it improves the precision from 68.8% to 97.8%. With the additional temporal prior volume, about 45% of the false positive metastases are reduced in the high sensitivity model and the precision reaches 99.6% for the high specificity model. The mean dice coefficient for all metastases is about 0.81. With the ensemble of the high sensitivity and high specificity models, on average only 1.5 false positive metastases per patient needs further check, while the majority of true positive metastases are confirmed. The ensemble learning is able to distinguish high confidence true positive metastases from metastases candidates that require special expert review or further follow-up, being particularly well-fit to the requirements of expert support in real clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge