Stefan Fischer
Deep Learning for automatic head and neck lymph node level delineation
Aug 28, 2022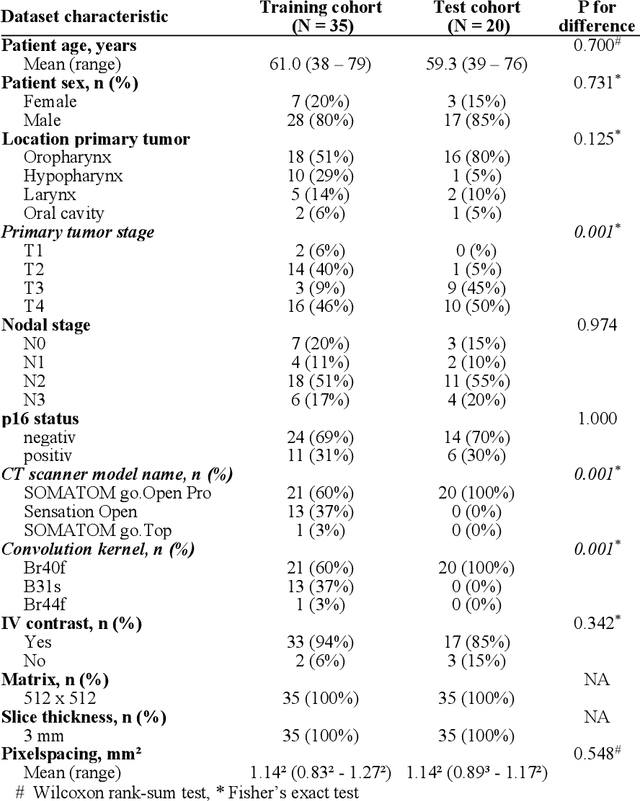
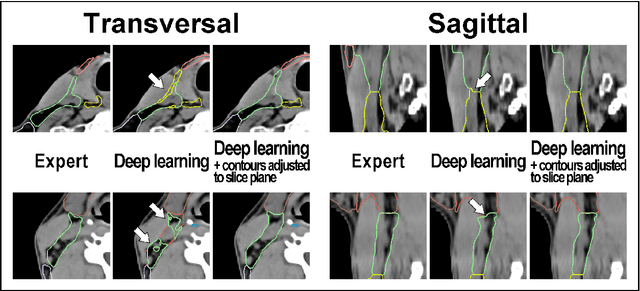
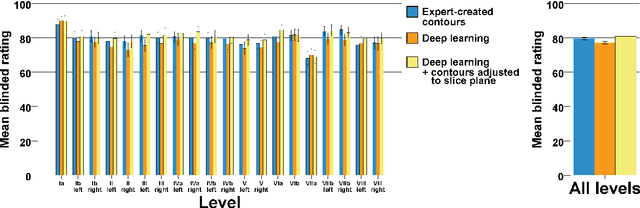
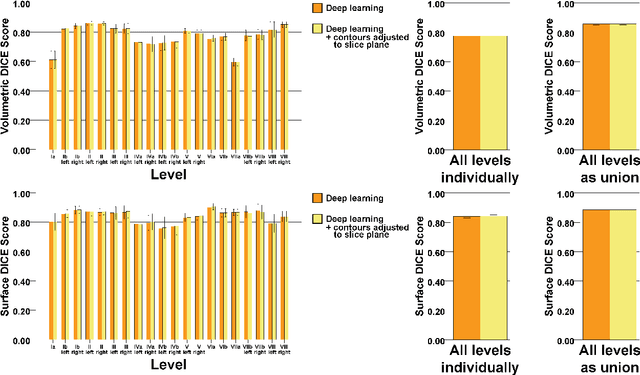
Abstract:Background: Deep learning-based head and neck lymph node level (HN_LNL) autodelineation is of high relevance to radiotherapy research and clinical treatment planning but still understudied in academic literature. Methods: An expert-delineated cohort of 35 planning CTs was used for training of an nnU-net 3D-fullres/2D-ensemble model for autosegmentation of 20 different HN_LNL. Validation was performed in an independent test set (n=20). In a completely blinded evaluation, 3 clinical experts rated the quality of deep learning autosegmentations in a head-to-head comparison with expert-created contours. For a subgroup of 10 cases, intraobserver variability was compared to deep learning autosegmentation performance. The effect of autocontour consistency with CT slice plane orientation on geometric accuracy and expert rating was investigated. Results: Mean blinded expert rating per level was significantly better for deep learning segmentations with CT slice plane adjustment than for expert-created contours (81.0 vs. 79.6, p<0.001), but deep learning segmentations without slice plane adjustment were rated significantly worse than expert-created contours (77.2 vs. 79.6, p<0.001). Geometric accuracy of deep learning segmentations was non-different from intraobserver variability (mean Dice per level, 0.78 vs. 0.77, p=0.064) with variance in accuracy between levels being improved (p<0.001). Clinical significance of contour consistency with CT slice plane orientation was not represented by geometric accuracy metrics (Dice, 0.78 vs. 0.78, p=0.572) Conclusions: We show that a nnU-net 3D-fullres/2D-ensemble model can be used for highly accurate autodelineation of HN_LNL using only a limited training dataset that is ideally suited for large-scale standardized autodelineation of HN_LNL in the research setting. Geometric accuracy metrics are only an imperfect surrogate for blinded expert rating.
Continual Learning for Peer-to-Peer Federated Learning: A Study on Automated Brain Metastasis Identification
Apr 30, 2022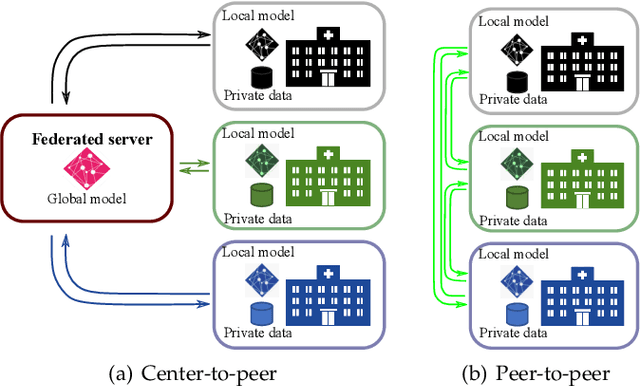
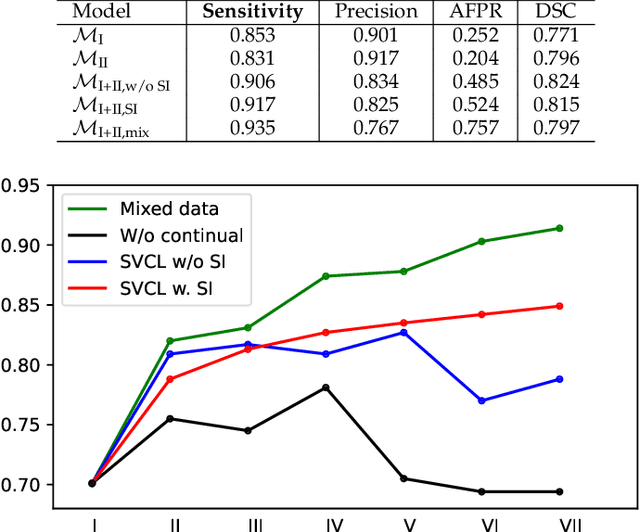
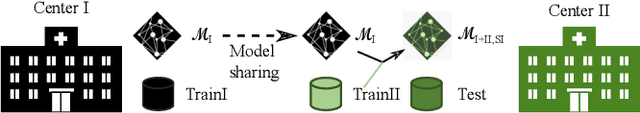
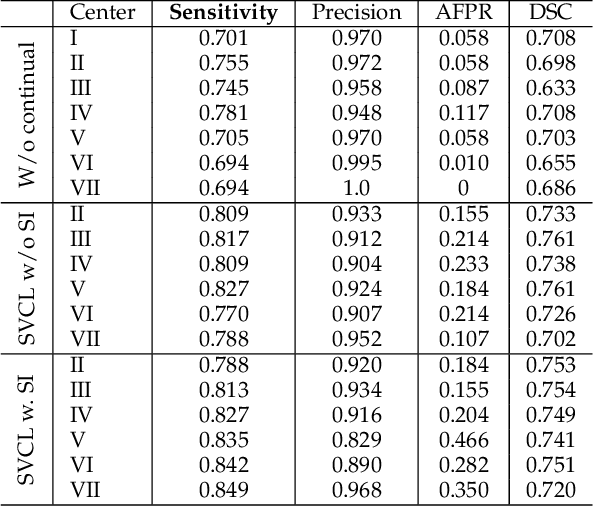
Abstract:Due to data privacy constraints, data sharing among multiple centers is restricted. Continual learning, as one approach to peer-to-peer federated learning, can promote multicenter collaboration on deep learning algorithm development by sharing intermediate models instead of training data. This work aims to investigate the feasibility of continual learning for multicenter collaboration on an exemplary application of brain metastasis identification using DeepMedic. 920 T1 MRI contrast enhanced volumes are split to simulate multicenter collaboration scenarios. A continual learning algorithm, synaptic intelligence (SI), is applied to preserve important model weights for training one center after another. In a bilateral collaboration scenario, continual learning with SI achieves a sensitivity of 0.917, and naive continual learning without SI achieves a sensitivity of 0.906, while two models trained on internal data solely without continual learning achieve sensitivity of 0.853 and 0.831 only. In a seven-center multilateral collaboration scenario, the models trained on internal datasets (100 volumes each center) without continual learning obtain a mean sensitivity value of 0.699. With single-visit continual learning (i.e., the shared model visits each center only once during training), the sensitivity is improved to 0.788 and 0.849 without SI and with SI, respectively. With iterative continual learning (i.e., the shared model revisits each center multiple times during training), the sensitivity is further improved to 0.914, which is identical to the sensitivity using mixed data for training. Our experiments demonstrate that continual learning can improve brain metastasis identification performance for centers with limited data. This study demonstrates the feasibility of applying continual learning for peer-to-peer federated learning in multicenter collaboration.
Lifting DecPOMDPs for Nanoscale Systems -- A Work in Progress
Oct 18, 2021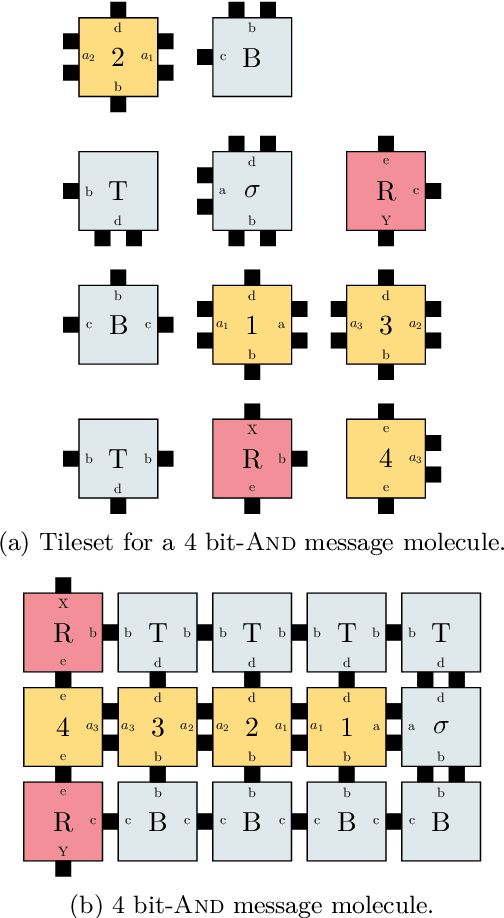
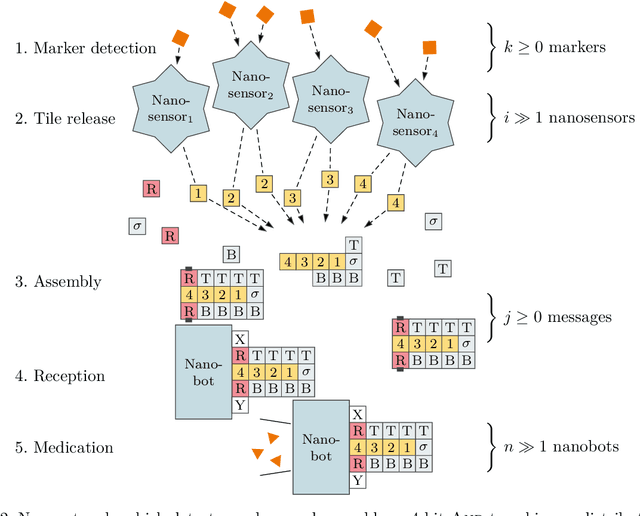
Abstract:DNA-based nanonetworks have a wide range of promising use cases, especially in the field of medicine. With a large set of agents, a partially observable stochastic environment, and noisy observations, such nanoscale systems can be modelled as a decentralised, partially observable, Markov decision process (DecPOMDP). As the agent set is a dominating factor, this paper presents (i) lifted DecPOMDPs, partitioning the agent set into sets of indistinguishable agents, reducing the worst-case space required, and (ii) a nanoscale medical system as an application. Future work turns to solving and implementing lifted DecPOMDPs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge