Arnd Dörfler
A Self-supervised Multimodal Deep Learning Approach to Differentiate Post-radiotherapy Progression from Pseudoprogression in Glioblastoma
Feb 06, 2025



Abstract:Accurate differentiation of pseudoprogression (PsP) from True Progression (TP) following radiotherapy (RT) in glioblastoma (GBM) patients is crucial for optimal treatment planning. However, this task remains challenging due to the overlapping imaging characteristics of PsP and TP. This study therefore proposes a multimodal deep-learning approach utilizing complementary information from routine anatomical MR images, clinical parameters, and RT treatment planning information for improved predictive accuracy. The approach utilizes a self-supervised Vision Transformer (ViT) to encode multi-sequence MR brain volumes to effectively capture both global and local context from the high dimensional input. The encoder is trained in a self-supervised upstream task on unlabeled glioma MRI datasets from the open BraTS2021, UPenn-GBM, and UCSF-PDGM datasets to generate compact, clinically relevant representations from FLAIR and T1 post-contrast sequences. These encoded MR inputs are then integrated with clinical data and RT treatment planning information through guided cross-modal attention, improving progression classification accuracy. This work was developed using two datasets from different centers: the Burdenko Glioblastoma Progression Dataset (n = 59) for training and validation, and the GlioCMV progression dataset from the University Hospital Erlangen (UKER) (n = 20) for testing. The proposed method achieved an AUC of 75.3%, outperforming the current state-of-the-art data-driven approaches. Importantly, the proposed approach relies on readily available anatomical MRI sequences, clinical data, and RT treatment planning information, enhancing its clinical feasibility. The proposed approach addresses the challenge of limited data availability for PsP and TP differentiation and could allow for improved clinical decision-making and optimized treatment plans for GBM patients.
Comprehensive Multimodal Deep Learning Survival Prediction Enabled by a Transformer Architecture: A Multicenter Study in Glioblastoma
May 21, 2024Abstract:Background: This research aims to improve glioblastoma survival prediction by integrating MR images, clinical and molecular-pathologic data in a transformer-based deep learning model, addressing data heterogeneity and performance generalizability. Method: We propose and evaluate a transformer-based non-linear and non-proportional survival prediction model. The model employs self-supervised learning techniques to effectively encode the high-dimensional MRI input for integration with non-imaging data using cross-attention. To demonstrate model generalizability, the model is assessed with the time-dependent concordance index (Cdt) in two training setups using three independent public test sets: UPenn-GBM, UCSF-PDGM, and RHUH-GBM, each comprising 378, 366, and 36 cases, respectively. Results: The proposed transformer model achieved promising performance for imaging as well as non-imaging data, effectively integrating both modalities for enhanced performance (UPenn-GBM test-set, imaging Cdt 0.645, multimodal Cdt 0.707) while outperforming state-of-the-art late-fusion 3D-CNN-based models. Consistent performance was observed across the three independent multicenter test sets with Cdt values of 0.707 (UPenn-GBM, internal test set), 0.672 (UCSF-PDGM, first external test set) and 0.618 (RHUH-GBM, second external test set). The model achieved significant discrimination between patients with favorable and unfavorable survival for all three datasets (logrank p 1.9\times{10}^{-8}, 9.7\times{10}^{-3}, and 1.2\times{10}^{-2}). Conclusions: The proposed transformer-based survival prediction model integrates complementary information from diverse input modalities, contributing to improved glioblastoma survival prediction compared to state-of-the-art methods. Consistent performance was observed across institutions supporting model generalizability.
Risk Classification of Brain Metastases via Radiomics, Delta-Radiomics and Machine Learning
Feb 17, 2023



Abstract:Stereotactic radiotherapy (SRT) is one of the most important treatment for patients with brain metastases (BM). Conventionally, following SRT patients are monitored by serial imaging and receive salvage treatments in case of significant tumor growth. We hypothesized that using radiomics and machine learning (ML), metastases at high risk for subsequent progression could be identified during follow-up prior to the onset of significant tumor growth, enabling personalized follow-up intervals and early selection for salvage treatment. All experiments are performed on a dataset from clinical routine of the Radiation Oncology department of the University Hospital Erlangen (UKER). The classification is realized via the maximum-relevance minimal-redundancy (MRMR) technique and support vector machines (SVM). The pipeline leads to a classification with a mean area under the curve (AUC) score of 0.83 in internal cross-validation and allows a division of the cohort into two subcohorts that differ significantly in their median time to progression (low-risk metastasis (LRM): 17.3 months, high-risk metastasis (HRM): 9.6 months, p < 0.01). The classification performance is especially enhanced by the analysis of medical images from different points in time (AUC 0.53 -> AUC 0.74). The results indicate that risk stratification of BM based on radiomics and machine learning during post-SRT follow-up is possible with good accuracy and should be further pursued to personalize and improve post-SRT follow-up.
snapshot CEST++ : the next snapshot CEST for fast whole-brain APTw imaging at 3T
Jul 01, 2022
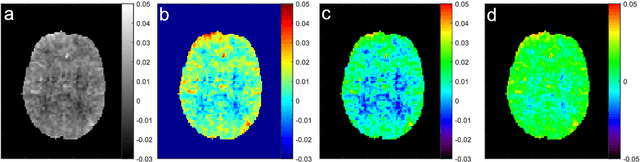
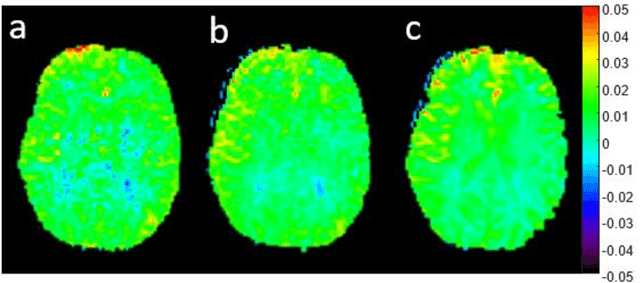
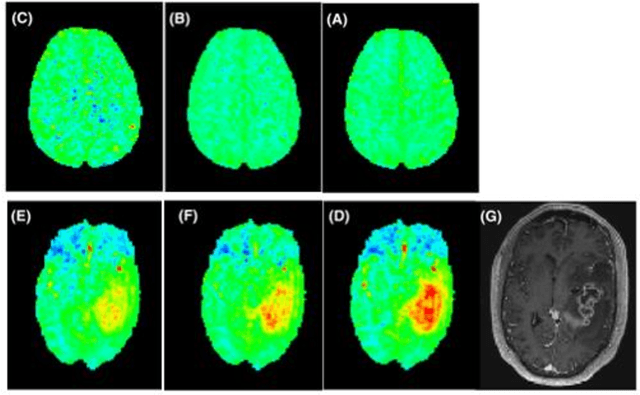
Abstract:CEST suffers from two main problems long acquisitin times or restricted coverage as well as incoherent protocol settings. In this paper we give suggestions on how to optimise your protocol settings fro CEST and present one setting for APT CEST. To increase the coverage while keeping the acquisition time constant we suggest using a spatial temporal Compressed Sensing approach. Finally, 1.8mm isotropic whole brain APT CEST maps can be acquired in a little bit less than 2min with a fully integrated online reconstruction. This will pave the way to an even further clinical use of CEST.
Continual Learning for Peer-to-Peer Federated Learning: A Study on Automated Brain Metastasis Identification
Apr 30, 2022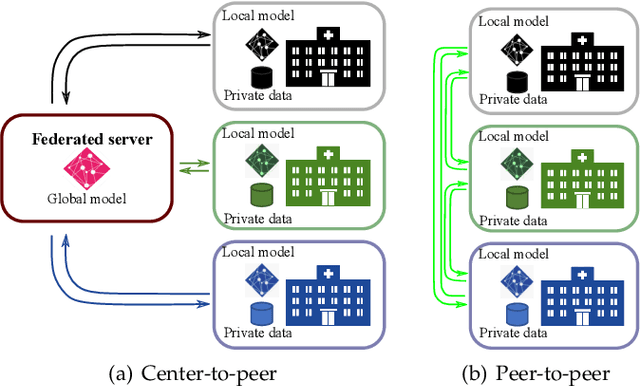
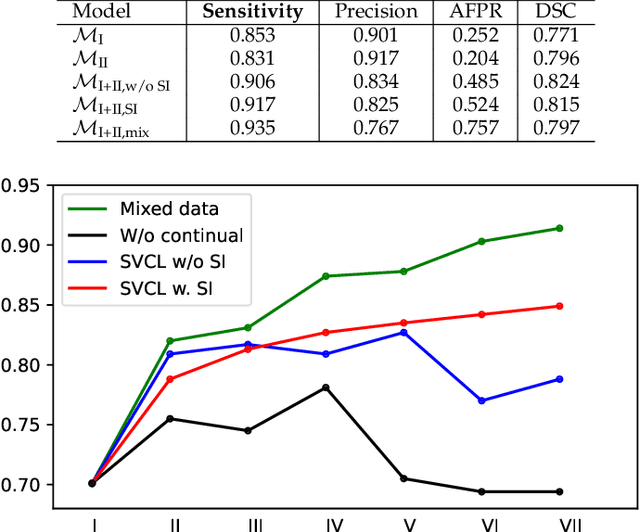
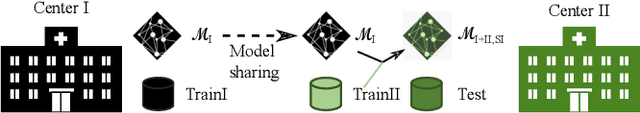
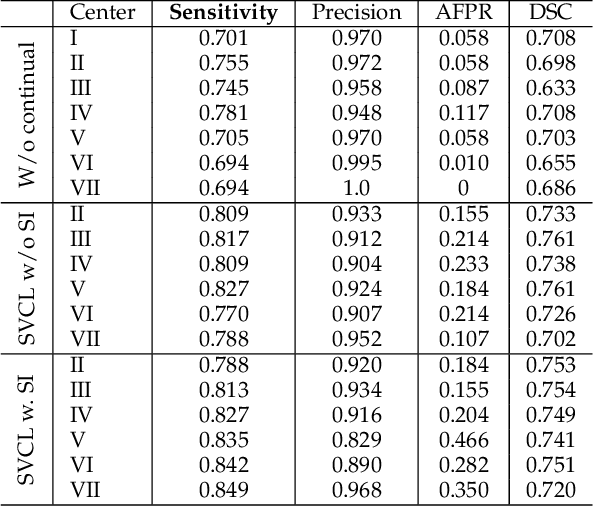
Abstract:Due to data privacy constraints, data sharing among multiple centers is restricted. Continual learning, as one approach to peer-to-peer federated learning, can promote multicenter collaboration on deep learning algorithm development by sharing intermediate models instead of training data. This work aims to investigate the feasibility of continual learning for multicenter collaboration on an exemplary application of brain metastasis identification using DeepMedic. 920 T1 MRI contrast enhanced volumes are split to simulate multicenter collaboration scenarios. A continual learning algorithm, synaptic intelligence (SI), is applied to preserve important model weights for training one center after another. In a bilateral collaboration scenario, continual learning with SI achieves a sensitivity of 0.917, and naive continual learning without SI achieves a sensitivity of 0.906, while two models trained on internal data solely without continual learning achieve sensitivity of 0.853 and 0.831 only. In a seven-center multilateral collaboration scenario, the models trained on internal datasets (100 volumes each center) without continual learning obtain a mean sensitivity value of 0.699. With single-visit continual learning (i.e., the shared model visits each center only once during training), the sensitivity is improved to 0.788 and 0.849 without SI and with SI, respectively. With iterative continual learning (i.e., the shared model revisits each center multiple times during training), the sensitivity is further improved to 0.914, which is identical to the sensitivity using mixed data for training. Our experiments demonstrate that continual learning can improve brain metastasis identification performance for centers with limited data. This study demonstrates the feasibility of applying continual learning for peer-to-peer federated learning in multicenter collaboration.
Deep learning for brain metastasis detection and segmentation in longitudinal MRI data
Dec 28, 2021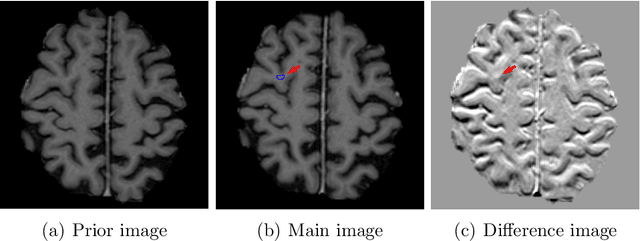
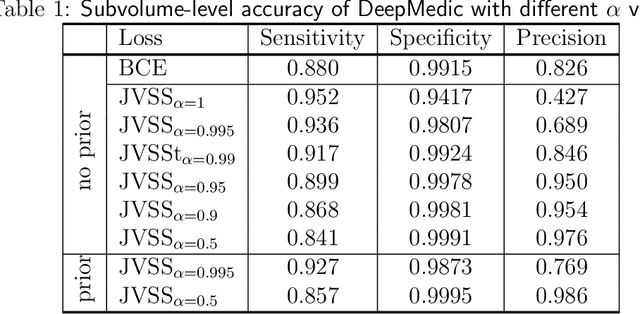
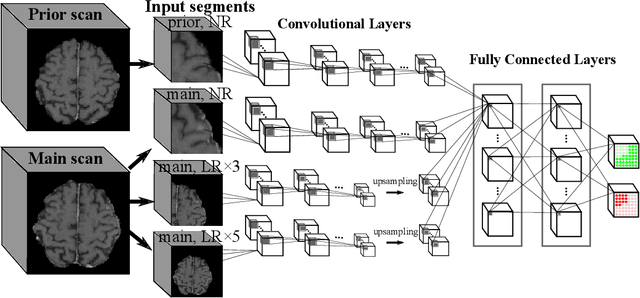
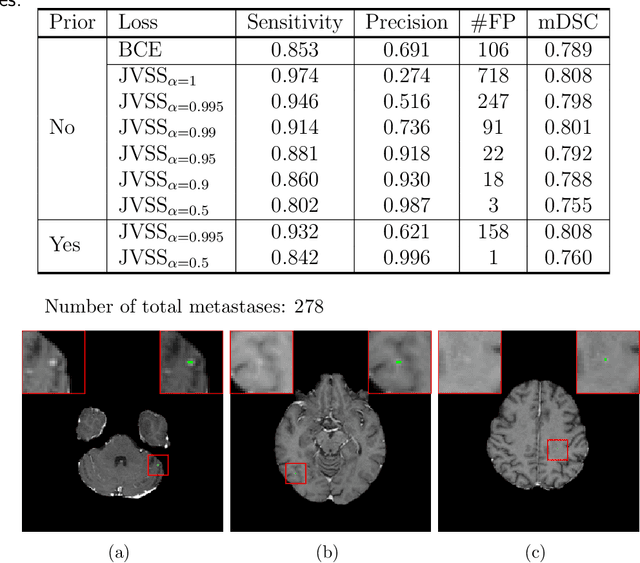
Abstract:Brain metastases occur frequently in patients with metastatic cancer. Early and accurate detection of brain metastases is very essential for treatment planning and prognosis in radiation therapy. To improve brain metastasis detection performance with deep learning, a custom detection loss called volume-level sensitivity-specificity (VSS) is proposed, which rates individual metastasis detection sensitivity and specificity in (sub-)volume levels. As sensitivity and precision are always a trade-off in a metastasis level, either a high sensitivity or a high precision can be achieved by adjusting the weights in the VSS loss without decline in dice score coefficient for segmented metastases. To reduce metastasis-like structures being detected as false positive metastases, a temporal prior volume is proposed as an additional input of the neural network. Our proposed VSS loss improves the sensitivity of brain metastasis detection, increasing the sensitivity from 86.7% to 95.5%. Alternatively, it improves the precision from 68.8% to 97.8%. With the additional temporal prior volume, about 45% of the false positive metastases are reduced in the high sensitivity model and the precision reaches 99.6% for the high specificity model. The mean dice coefficient for all metastases is about 0.81. With the ensemble of the high sensitivity and high specificity models, on average only 1.5 false positive metastases per patient needs further check, while the majority of true positive metastases are confirmed. The ensemble learning is able to distinguish high confidence true positive metastases from metastases candidates that require special expert review or further follow-up, being particularly well-fit to the requirements of expert support in real clinical practice.
Projection-to-Projection Translation for Hybrid X-ray and Magnetic Resonance Imaging
Nov 19, 2019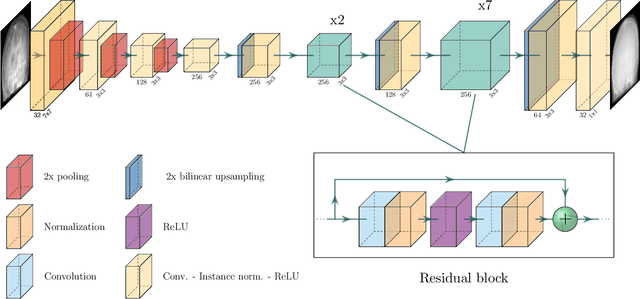
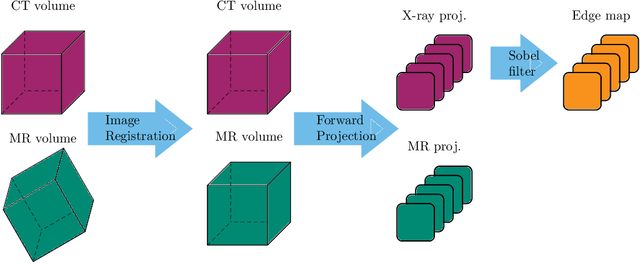
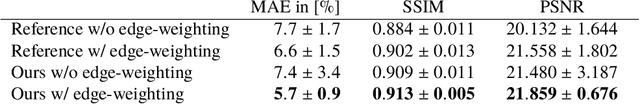
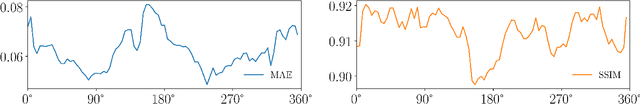
Abstract:Hybrid X-ray and magnetic resonance (MR) imaging promises large potential in interventional medical imaging applications due to the broad variety of contrast of MRI combined with fast imaging of X-ray-based modalities. To fully utilize the potential of the vast amount of existing image enhancement techniques, the corresponding information from both modalities must be present in the same domain. For image-guided interventional procedures, X-ray fluoroscopy has proven to be the modality of choice. Synthesizing one modality from another in this case is an ill-posed problem due to ambiguous signal and overlapping structures in projective geometry. To take on these challenges, we present a learning-based solution to MR to X-ray projection-to-projection translation. We propose an image generator network that focuses on high representation capacity in higher resolution layers to allow for accurate synthesis of fine details in the projection images. Additionally, a weighting scheme in the loss computation that favors high-frequency structures is proposed to focus on the important details and contours in projection imaging. The proposed extensions prove valuable in generating X-ray projection images with natural appearance. Our approach achieves a deviation from the ground truth of only $6$% and structural similarity measure of $0.913\,\pm\,0.005$. In particular the high frequency weighting assists in generating projection images with sharp appearance and reduces erroneously synthesized fine details.
Multi-modal Deep Guided Filtering for Comprehensible Medical Image Processing
Nov 18, 2019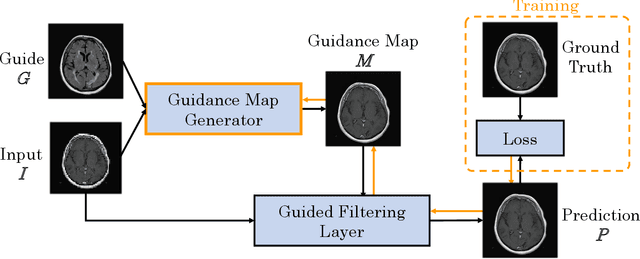
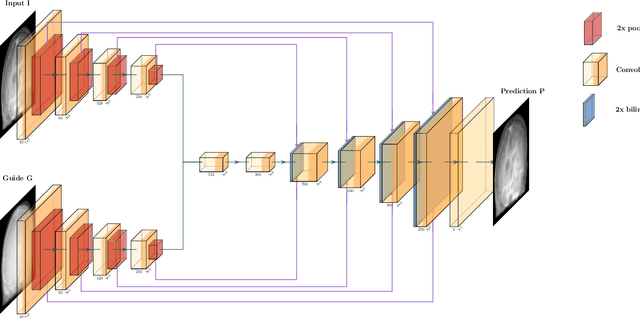
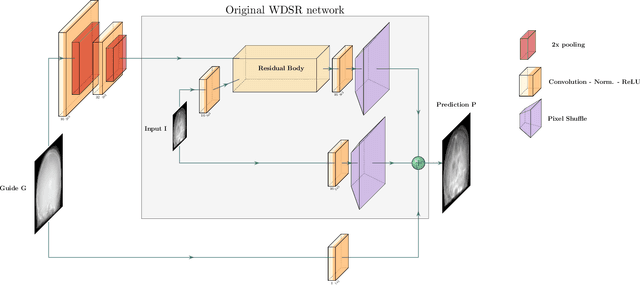
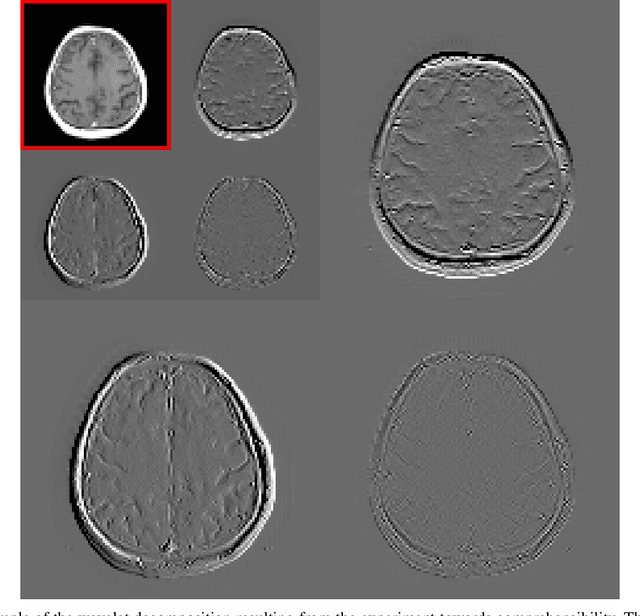
Abstract:Deep learning-based image processing is capable of creating highly appealing results. However, it is still widely considered as a "blackbox" transformation. In medical imaging, this lack of comprehensibility of the results is a sensitive issue. The integration of known operators into the deep learning environment has proven to be advantageous for the comprehensibility and reliability of the computations. Consequently, we propose the use of the locally linear guided filter in combination with a learned guidance map for general purpose medical image processing. The output images are only processed by the guided filter while the guidance map can be trained to be task-optimal in an end-to-end fashion. We investigate the performance based on two popular tasks: image super resolution and denoising. The evaluation is conducted based on pairs of multi-modal magnetic resonance imaging and cross-modal computed tomography and magnetic resonance imaging datasets. For both tasks, the proposed approach is on par with state-of-the-art approaches. Additionally, we can show that the input image's content is almost unchanged after the processing which is not the case for conventional deep learning approaches. On top, the proposed pipeline offers increased robustness against degraded input as well as adversarial attacks.
Deriving Neural Network Architectures using Precision Learning: Parallel-to-fan beam Conversion
Oct 23, 2018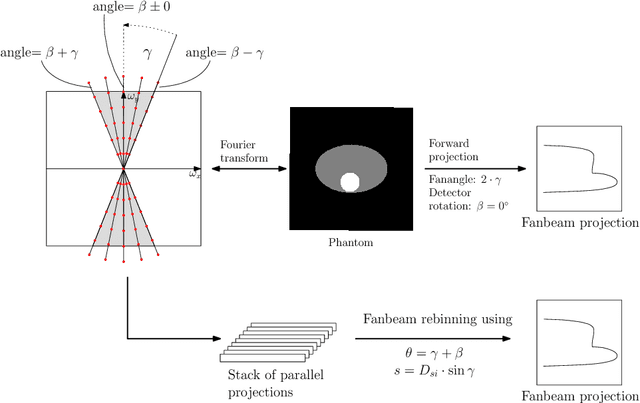
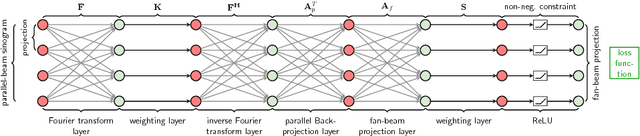
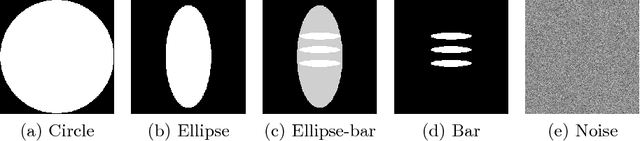
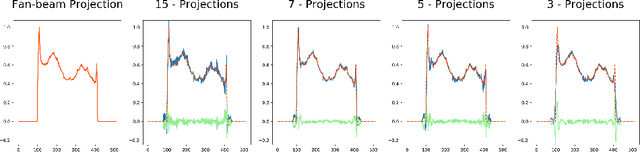
Abstract:In this paper, we derive a neural network architecture based on an analytical formulation of the parallel-to-fan beam conversion problem following the concept of precision learning. The network allows to learn the unknown operators in this conversion in a data-driven manner avoiding interpolation and potential loss of resolution. Integration of known operators results in a small number of trainable parameters that can be estimated from synthetic data only. The concept is evaluated in the context of Hybrid MRI/X-ray imaging where transformation of the parallel-beam MRI projections to fan-beam X-ray projections is required. The proposed method is compared to a traditional rebinning method. The results demonstrate that the proposed method is superior to ray-by-ray interpolation and is able to deliver sharper images using the same amount of parallel-beam input projections which is crucial for interventional applications. We believe that this approach forms a basis for further work uniting deep learning, signal processing, physics, and traditional pattern recognition.
Precision Learning: Reconstruction Filter Kernel Discretization
Jul 09, 2018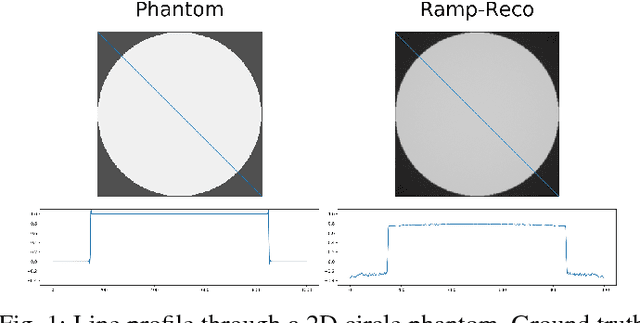
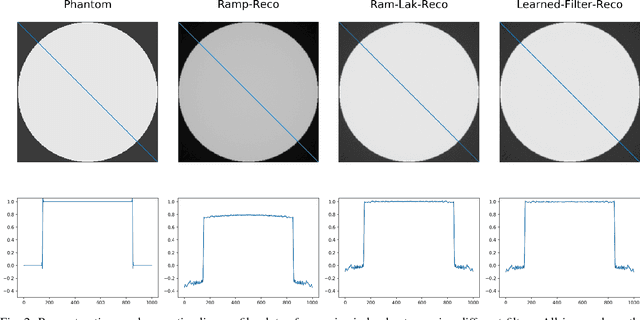
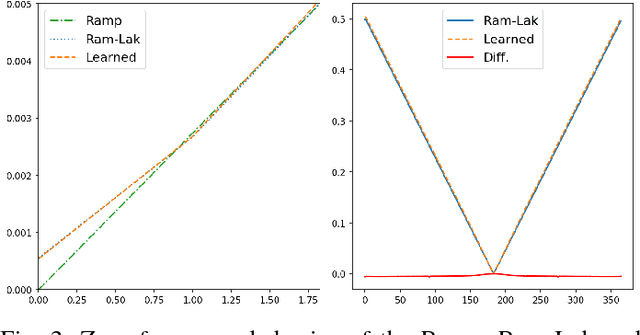
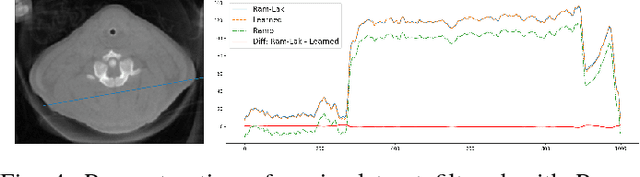
Abstract:In this paper, we present substantial evidence that a deep neural network will intrinsically learn the appropriate way to discretize the ideal continuous reconstruction filter. Currently, the Ram-Lak filter or heuristic filters which impose different noise assumptions are used for filtered back-projection. All of these, however, inhibit a fully data-driven reconstruction deep learning approach. In addition, the heuristic filters are not chosen in an optimal sense. To tackle this issue, we propose a formulation to directly learn the reconstruction filter. The filter is initialized with the ideal Ramp filter as a strong pre-training and learned in frequency domain. We compare the learned filter with the Ram-Lak and the Ramp filter on a numerical phantom as well as on a real CT dataset. The results show that the network properly discretizes the continuous Ramp filter and converges towards the Ram-Lak solution. In our view these observations are interesting to gain a better understanding of deep learning techniques and traditional analytic techniques such as Wiener filtering and discretization theory. Furthermore, this will allow fully trainable data-driven reconstruction deep learning approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge