Moritz Zaiss
Institute of Neuroradiology, Friedrich-Alexander Universitat Erlangen-Nurnberg, Department of Artificial Intelligence in Biomedical Engineering, Friedrich-Alexander Universitat Erlangen-Nurnberg, Erlangen, Germany
Multiparameter Uncertainty Mapping in Quantitative Molecular MRI using a Physics-Structured Variational Autoencoder (PS-VAE)
Feb 03, 2026Abstract:Quantitative imaging methods, such as magnetic resonance fingerprinting (MRF), aim to extract interpretable pathology biomarkers by estimating biophysical tissue parameters from signal evolutions. However, the pattern-matching algorithms or neural networks used in such inverse problems often lack principled uncertainty quantification, which limits the trustworthiness and transparency, required for clinical acceptance. Here, we describe a physics-structured variational autoencoder (PS-VAE) designed for rapid extraction of voxelwise multi-parameter posterior distributions. Our approach integrates a differentiable spin physics simulator with self-supervised learning, and provides a full covariance that captures the inter-parameter correlations of the latent biophysical space. The method was validated in a multi-proton pool chemical exchange saturation transfer (CEST) and semisolid magnetization transfer (MT) molecular MRF study, across in-vitro phantoms, tumor-bearing mice, healthy human volunteers, and a subject with glioblastoma. The resulting multi-parametric posteriors are in good agreement with those calculated using a brute-force Bayesian analysis, while providing an orders-of-magnitude acceleration in whole brain quantification. In addition, we demonstrate how monitoring the multi-parameter posterior dynamics across progressively acquired signals provides practical insights for protocol optimization and may facilitate real-time adaptive acquisition.
Multi-Parameter Molecular MRI Quantification using Physics-Informed Self-Supervised Learning
Nov 10, 2024Abstract:Biophysical model fitting plays a key role in obtaining quantitative parameters from physiological signals and images. However, the model complexity for molecular magnetic resonance imaging (MRI) often translates into excessive computation time, which makes clinical use impractical. Here, we present a generic computational approach for solving the parameter extraction inverse problem posed by ordinary differential equation (ODE) modeling coupled with experimental measurement of the system dynamics. This is achieved by formulating a numerical ODE solver to function as a step-wise analytical one, thereby making it compatible with automatic differentiation-based optimization. This enables efficient gradient-based model fitting, and provides a new approach to parameter quantification based on self-supervised learning from a single data observation. The neural-network-based train-by-fit pipeline was used to quantify semisolid magnetization transfer (MT) and chemical exchange saturation transfer (CEST) amide proton exchange parameters in the human brain, in an in-vivo molecular MRI study (n=4). The entire pipeline of the first whole brain quantification was completed in 18.3$\pm$8.3 minutes, which is an order-of-magnitude faster than comparable alternatives. Reusing the single-subject-trained network for inference in new subjects took 1.0$\pm$0.2 s, to provide results in agreement with literature values and scan-specific fit results (Pearson's r>0.98, p<0.0001).
Controlling sharpness, SNR and SAR for 3D FSE at 7T by end-to-end learning
Sep 30, 2024Abstract:Purpose: To non-heuristically identify dedicated variable flip angle (VFA) schemes optimized for the point-spread function (PSF) and signal-to-noise ratio (SNR) of multiple tissues in 3D FSE sequences with very long echo trains at 7T. Methods: The proposed optimization considers predefined SAR constraints and target contrast using an end-to-end learning framework. The cost function integrates components for contrast fidelity (SNR) and a penalty term to minimize image blurring (PSF) for multiple tissues. By adjusting the weights of PSF/SNR cost-function components, PSF- and SNR-optimized VFAs were derived and tested in vivo using both the open-source Pulseq standard on two volunteers as well as vendor protocols on a 7T MRI system with parallel transmit extension on three volunteers. Results: PSF-optimized VFAs resulted in significantly reduced image blurring compared to standard VFAs for T2w while maintaining contrast fidelity. Small white and gray matter structures, as well as blood vessels, are more visible with PSF-optimized VFAs. Quantitative analysis shows that the optimized VFA yields 50% less deviation from a sinc-like reference PSF than the standard VFA. The SNR-optimized VFAs yielded images with significantly improved SNR in a white and gray matter region relative to standard (81.2\pm18.4 vs. 41.2\pm11.5, respectively) as trade-off for elevated image blurring. Conclusion: This study demonstrates the potential of end-to-end learning frameworks to optimize VFA schemes in very long echo trains for 3D FSE acquisition at 7T in terms of PSF and SNR. It paves the way for fast and flexible adjustment of the trade-off between PSF and SNR for 3D FSE.
Decoding the human brain tissue response to radiofrequency excitation using a biophysical-model-free deep MRI on a chip framework
Aug 15, 2024



Abstract:Magnetic resonance imaging (MRI) relies on radiofrequency (RF) excitation of proton spin. Clinical diagnosis requires a comprehensive collation of biophysical data via multiple MRI contrasts, acquired using a series of RF sequences that lead to lengthy examinations. Here, we developed a vision transformer-based framework that captures the spatiotemporal magnetic signal evolution and decodes the brain tissue response to RF excitation, constituting an MRI on a chip. Following a per-subject rapid calibration scan (28.2 s), a wide variety of image contrasts including fully quantitative molecular, water relaxation, and magnetic field maps can be generated automatically. The method was validated across healthy subjects and a cancer patient in two different imaging sites, and proved to be 94% faster than alternative protocols. The deep MRI on a chip (DeepMonC) framework may reveal the molecular composition of the human brain tissue in a wide range of pathologies, while offering clinically attractive scan times.
BMapOpt: Optimization of Brain Tissue Probability Maps using a Differentiable MRI Simulator
Apr 23, 2024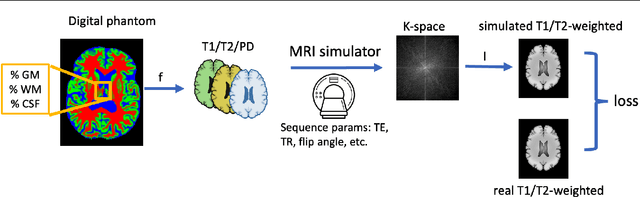
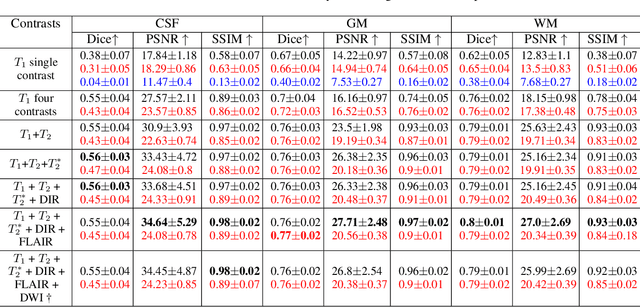
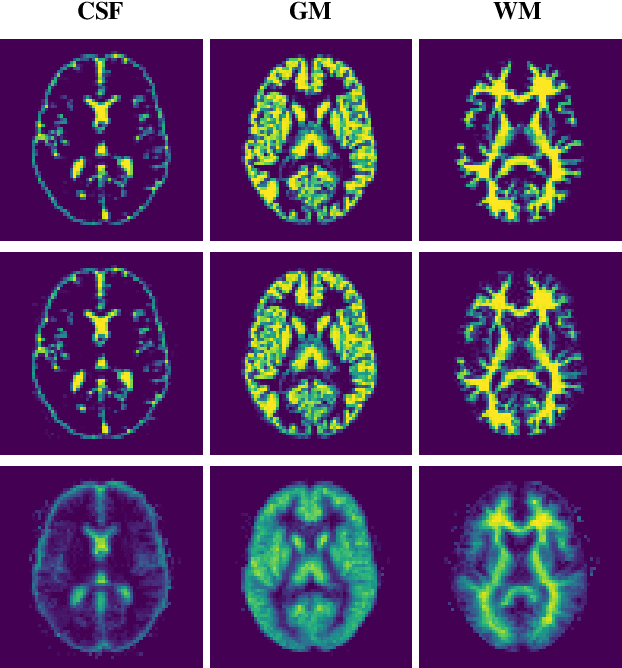
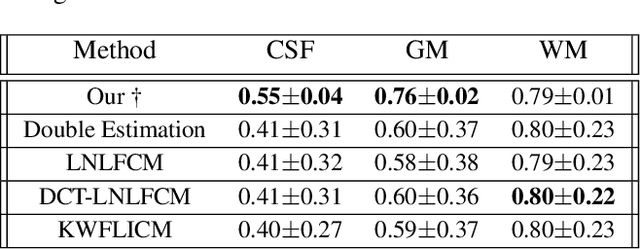
Abstract:Reconstructing digital brain phantoms in the form of multi-channeled brain tissue probability maps for individual subjects is essential for capturing brain anatomical variability, understanding neurological diseases, as well as for testing image processing methods. We demonstrate the first framework that optimizes brain tissue probability maps (Gray Matter - GM, White Matter - WM, and Cerebrospinal fluid - CSF) with the help of a Physics-based differentiable MRI simulator that models the magnetization signal at each voxel in the image. Given an observed $T_1$/$T_2$-weighted MRI scan, the corresponding clinical MRI sequence, and the MRI differentiable simulator, we optimize the simulator's input probability maps by back-propagating the L2 loss between the simulator's output and the $T_1$/$T_2$-weighted scan. This approach has the significant advantage of not relying on any training data, and instead uses the strong inductive bias of the MRI simulator. We tested the model on 20 scans from the BrainWeb database and demonstrate a highly accurate reconstruction of GM, WM, and CSF.
A novel image space formalism of Fourier domain interpolation neural networks for noise propagation analysis
Feb 27, 2024Abstract:Purpose: To develop an image space formalism of multi-layer convolutional neural networks (CNNs) for Fourier domain interpolation in MRI reconstructions and analytically estimate noise propagation during CNN inference. Theory and Methods: Nonlinear activations in the Fourier domain (also known as k-space) using complex-valued Rectifier Linear Units are expressed as elementwise multiplication with activation masks. This operation is transformed into a convolution in the image space. After network training in k-space, this approach provides an algebraic expression for the derivative of the reconstructed image with respect to the aliased coil images, which serve as the input tensors to the network in the image space. This allows the variance in the network inference to be estimated analytically and to be used to describe noise characteristics. Monte-Carlo simulations and numerical approaches based on auto-differentiation were used for validation. The framework was tested on retrospectively undersampled invivo brain images. Results: Inferences conducted in the image domain are quasi-identical to inferences in the k-space, underlined by corresponding quantitative metrics. Noise variance maps obtained from the analytical expression correspond with those obtained via Monte-Carlo simulations, as well as via an auto-differentiation approach. The noise resilience is well characterized, as in the case of classical Parallel Imaging. Komolgorov-Smirnov tests demonstrate Gaussian distributions of voxel magnitudes in variance maps obtained via Monte-Carlo simulations. Conclusion: The quasi-equivalent image space formalism for neural networks for k-space interpolation enables fast and accurate description of the noise characteristics during CNN inference, analogous to geometry-factor maps in traditional parallel imaging methods.
Joint MR sequence optimization beats pure neural network approaches for spin-echo MRI super-resolution
May 12, 2023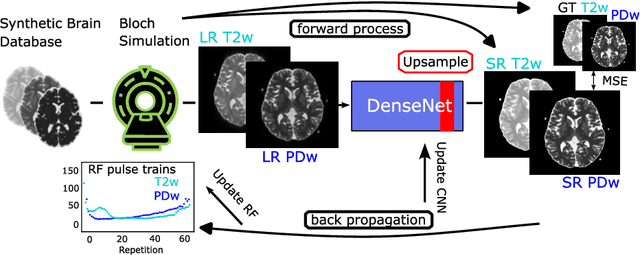
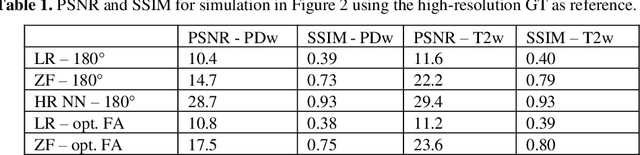
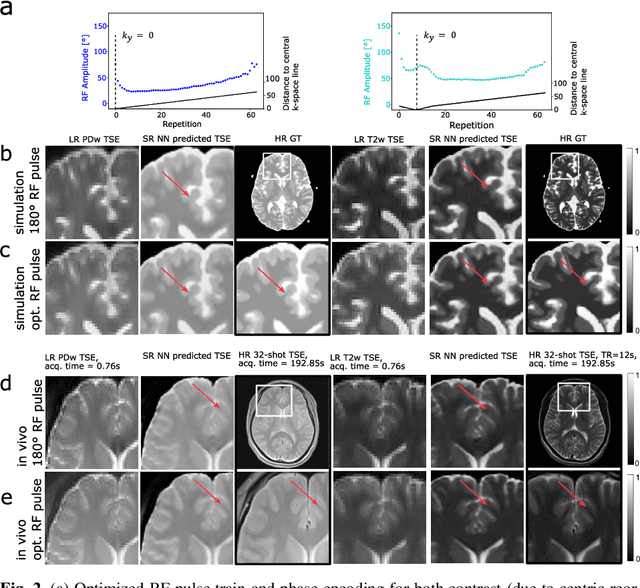
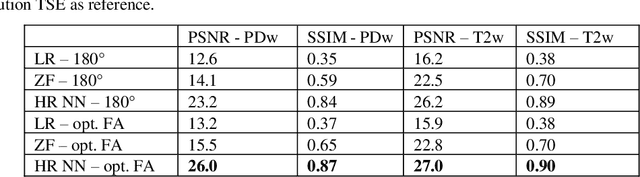
Abstract:Current MRI super-resolution (SR) methods only use existing contrasts acquired from typical clinical sequences as input for the neural network (NN). In turbo spin echo sequences (TSE) the sequence parameters can have a strong influence on the actual resolution of the acquired image and have consequently a considera-ble impact on the performance of the NN. We propose a known-operator learning approach to perform an end-to-end optimization of MR sequence and neural net-work parameters for SR-TSE. This MR-physics-informed training procedure jointly optimizes the radiofrequency pulse train of a proton density- (PD-) and T2-weighted TSE and a subsequently applied convolutional neural network to predict the corresponding PDw and T2w super-resolution TSE images. The found radiofrequency pulse train designs generate an optimal signal for the NN to perform the SR task. Our method generalizes from the simulation-based optimi-zation to in vivo measurements and the acquired physics-informed SR images show higher correlation with a time-consuming segmented high-resolution TSE sequence compared to a pure network training approach.
Scale-Equivariant Deep Learning for 3D Data
Apr 12, 2023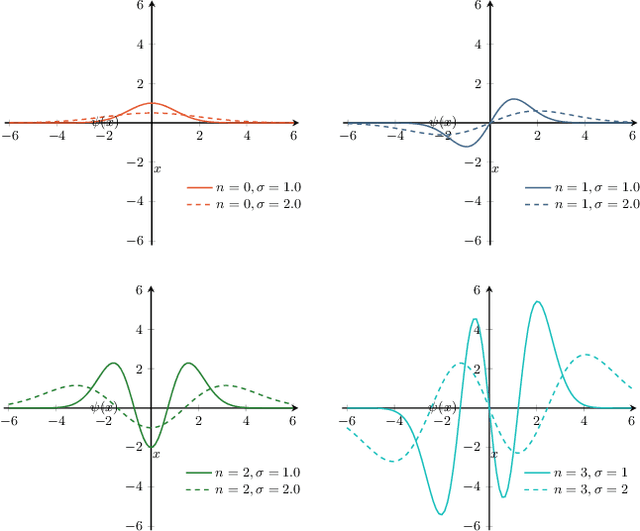
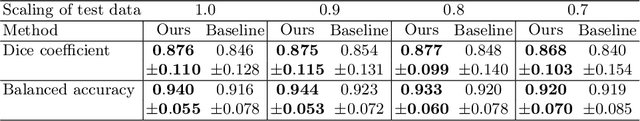
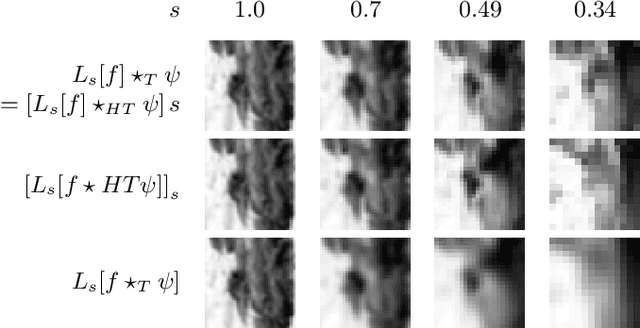
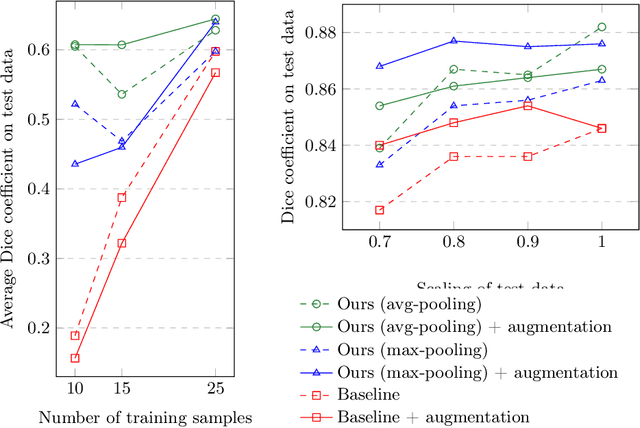
Abstract:The ability of convolutional neural networks (CNNs) to recognize objects regardless of their position in the image is due to the translation-equivariance of the convolutional operation. Group-equivariant CNNs transfer this equivariance to other transformations of the input. Dealing appropriately with objects and object parts of different scale is challenging, and scale can vary for multiple reasons such as the underlying object size or the resolution of the imaging modality. In this paper, we propose a scale-equivariant convolutional network layer for three-dimensional data that guarantees scale-equivariance in 3D CNNs. Scale-equivariance lifts the burden of having to learn each possible scale separately, allowing the neural network to focus on higher-level learning goals, which leads to better results and better data-efficiency. We provide an overview of the theoretical foundations and scientific work on scale-equivariant neural networks in the two-dimensional domain. We then transfer the concepts from 2D to the three-dimensional space and create a scale-equivariant convolutional layer for 3D data. Using the proposed scale-equivariant layer, we create a scale-equivariant U-Net for medical image segmentation and compare it with a non-scale-equivariant baseline method. Our experiments demonstrate the effectiveness of the proposed method in achieving scale-equivariance for 3D medical image analysis. We publish our code at https://github.com/wimmerth/scale-equivariant-3d-convnet for further research and application.
Accelerated and Quantitative 3D Semisolid MT/CEST Imaging using a Generative Adversarial Network (GAN-CEST)
Jul 22, 2022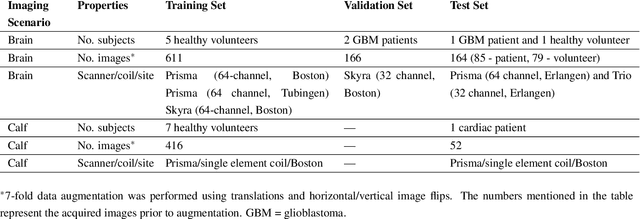
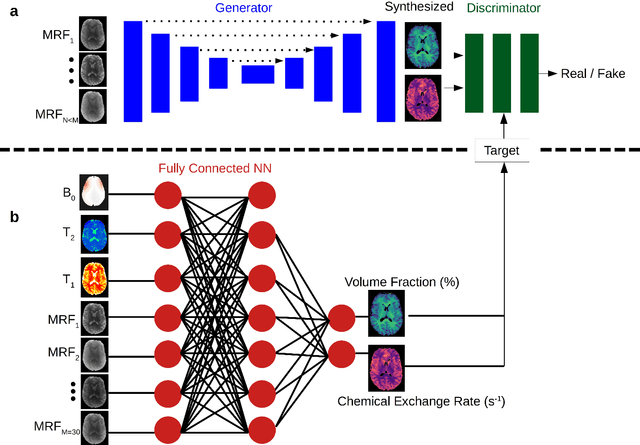
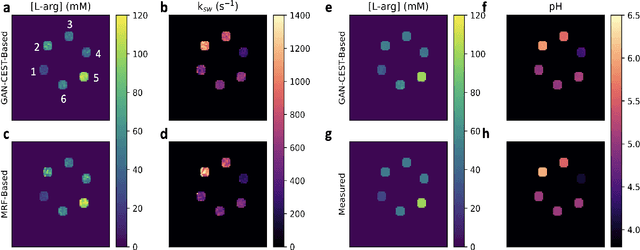
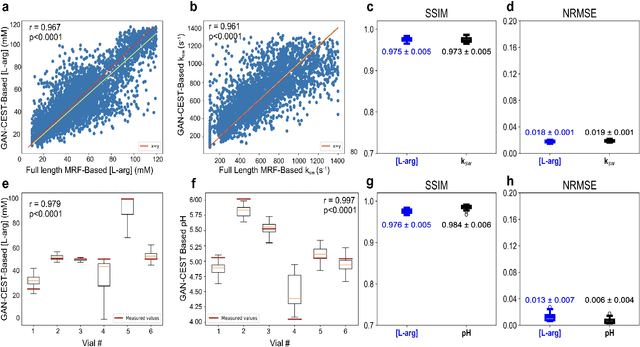
Abstract:Purpose: To substantially shorten the acquisition time required for quantitative 3D chemical exchange saturation transfer (CEST) and semisolid magnetization transfer (MT) imaging and allow for rapid chemical exchange parameter map reconstruction. Methods: Three-dimensional CEST and MT magnetic resonance fingerprinting (MRF) datasets of L-arginine phantoms, whole-brains, and calf muscles from healthy volunteers, cancer patients, and cardiac patients were acquired using 3T clinical scanners at 3 different sites, using 3 different scanner models and coils. A generative adversarial network supervised framework (GAN-CEST) was then designed and trained to learn the mapping from a reduced input data space to the quantitative exchange parameter space, while preserving perceptual and quantitative content. Results: The GAN-CEST 3D acquisition time was 42-52 seconds, 70% shorter than CEST-MRF. The quantitative reconstruction of the entire brain took 0.8 seconds. An excellent agreement was observed between the ground truth and GAN-based L-arginine concentration and pH values (Pearson's r > 0.97, NRMSE < 1.5%). GAN-CEST images from a brain-tumor subject yielded a semi-solid volume fraction and exchange rate NRMSE of 3.8$\pm$1.3% and 4.6$\pm$1.3%, respectively, and SSIM of 96.3$\pm$1.6% and 95.0$\pm$2.4%, respectively. The mapping of the calf-muscle exchange parameters in a cardiac patient, yielded NRMSE < 7% and SSIM > 94% for the semi-solid exchange parameters. In regions with large susceptibility artifacts, GAN-CEST has demonstrated improved performance and reduced noise compared to MRF. Conclusion: GAN-CEST can substantially reduce the acquisition time for quantitative semisolid MT/CEST mapping, while retaining performance even when facing pathologies and scanner models that were not available during training.
snapshot CEST++ : the next snapshot CEST for fast whole-brain APTw imaging at 3T
Jul 01, 2022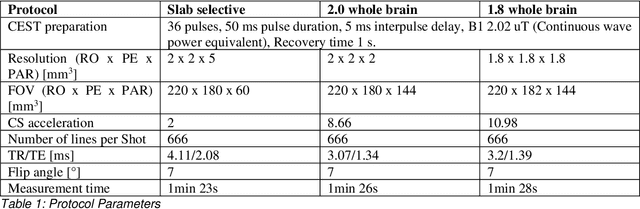
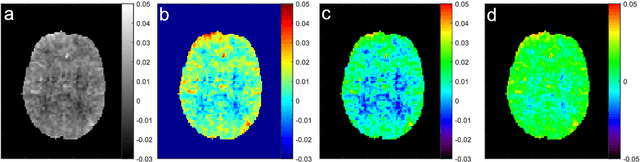
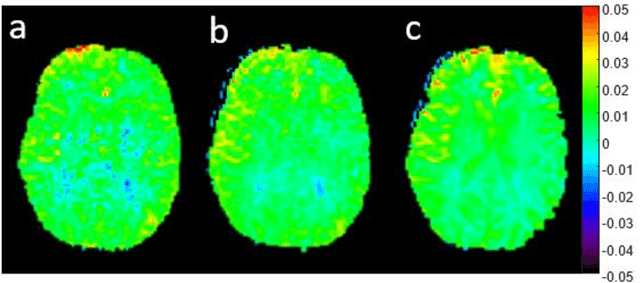
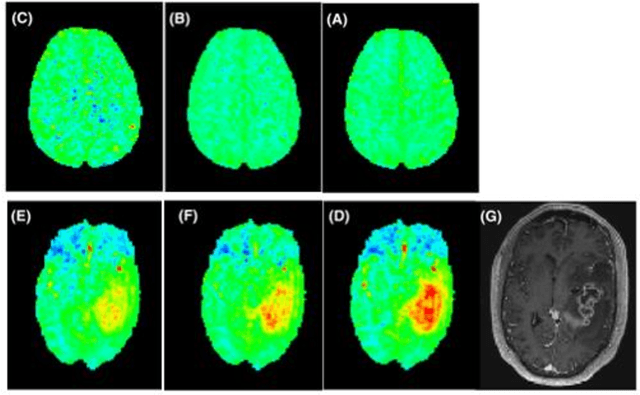
Abstract:CEST suffers from two main problems long acquisitin times or restricted coverage as well as incoherent protocol settings. In this paper we give suggestions on how to optimise your protocol settings fro CEST and present one setting for APT CEST. To increase the coverage while keeping the acquisition time constant we suggest using a spatial temporal Compressed Sensing approach. Finally, 1.8mm isotropic whole brain APT CEST maps can be acquired in a little bit less than 2min with a fully integrated online reconstruction. This will pave the way to an even further clinical use of CEST.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge