Vladimir Golkov
Generation of Musical Timbres using a Text-Guided Diffusion Model
Apr 12, 2025Abstract:In recent years, text-to-audio systems have achieved remarkable success, enabling the generation of complete audio segments directly from text descriptions. While these systems also facilitate music creation, the element of human creativity and deliberate expression is often limited. In contrast, the present work allows composers, arrangers, and performers to create the basic building blocks for music creation: audio of individual musical notes for use in electronic instruments and DAWs. Through text prompts, the user can specify the timbre characteristics of the audio. We introduce a system that combines a latent diffusion model and multi-modal contrastive learning to generate musical timbres conditioned on text descriptions. By jointly generating the magnitude and phase of the spectrogram, our method eliminates the need for subsequently running a phase retrieval algorithm, as related methods do. Audio examples, source code, and a web app are available at https://wxuanyuan.github.io/Musical-Note-Generation/
Interactions Across Blocks in Post-Training Quantization of Large Language Models
Nov 06, 2024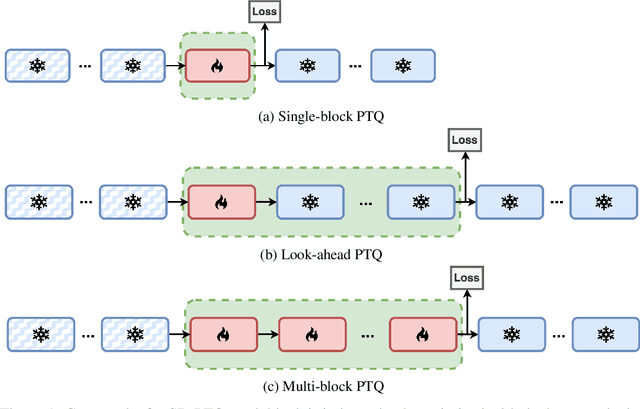
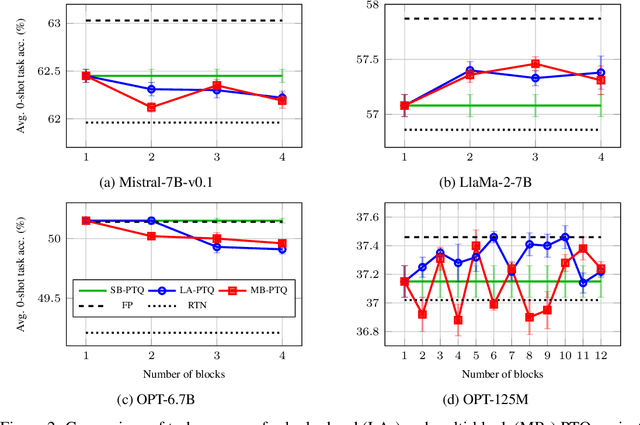
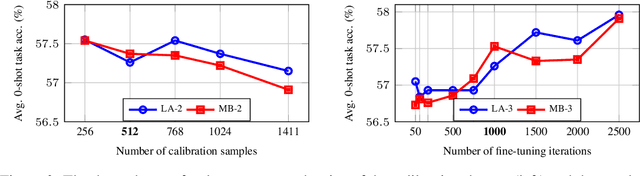

Abstract:Post-training quantization is widely employed to reduce the computational demands of neural networks. Typically, individual substructures, such as layers or blocks of layers, are quantized with the objective of minimizing quantization errors in their pre-activations by fine-tuning the corresponding weights. Deriving this local objective from the global objective of minimizing task loss involves two key simplifications: assuming substructures are mutually independent and ignoring the knowledge of subsequent substructures as well as the task loss. In this work, we assess the effects of these simplifications on weight-only quantization of large language models. We introduce two multi-block fine-tuning strategies and compare them against the baseline of fine-tuning single transformer blocks. The first captures correlations of weights across blocks by jointly optimizing multiple quantized blocks. The second incorporates knowledge of subsequent blocks by minimizing the error in downstream pre-activations rather than focusing solely on the quantized block. Our findings indicate that the effectiveness of these methods depends on the specific network model, with no impact on some models but demonstrating significant benefits for others.
How to Choose a Reinforcement-Learning Algorithm
Jul 30, 2024
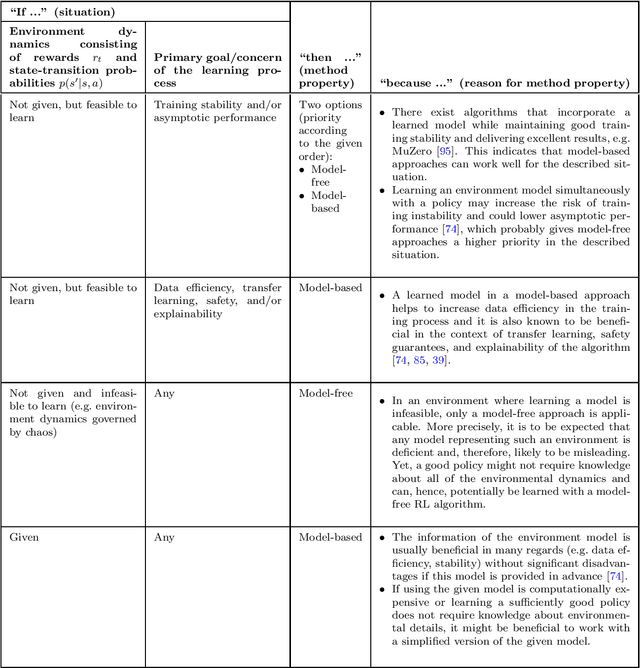
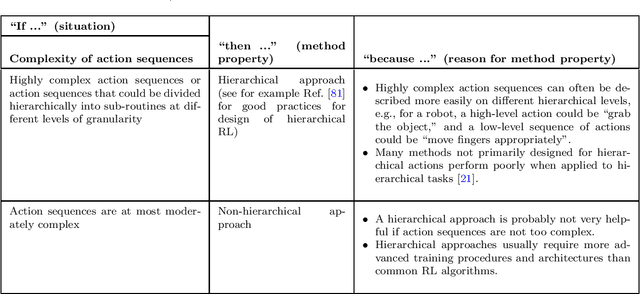

Abstract:The field of reinforcement learning offers a large variety of concepts and methods to tackle sequential decision-making problems. This variety has become so large that choosing an algorithm for a task at hand can be challenging. In this work, we streamline the process of choosing reinforcement-learning algorithms and action-distribution families. We provide a structured overview of existing methods and their properties, as well as guidelines for when to choose which methods. An interactive version of these guidelines is available online at https://rl-picker.github.io/.
Joint MR sequence optimization beats pure neural network approaches for spin-echo MRI super-resolution
May 12, 2023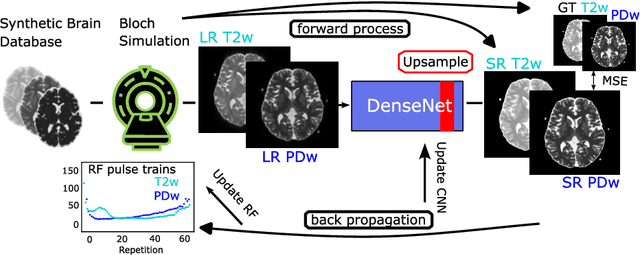
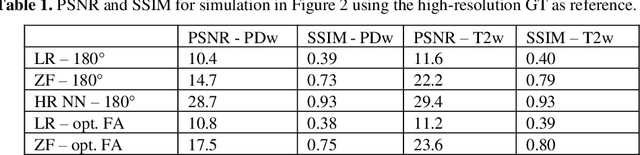
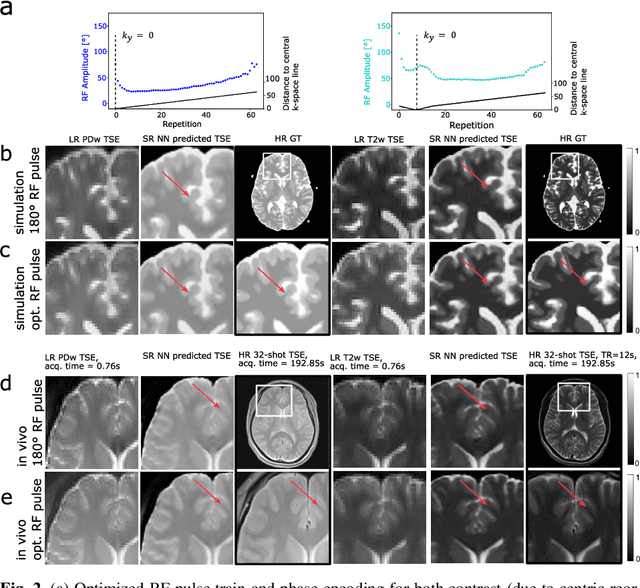
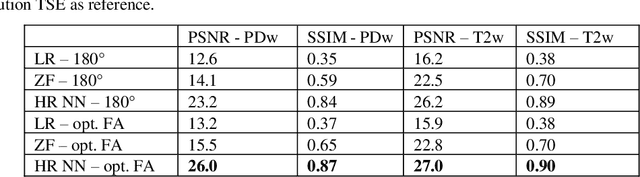
Abstract:Current MRI super-resolution (SR) methods only use existing contrasts acquired from typical clinical sequences as input for the neural network (NN). In turbo spin echo sequences (TSE) the sequence parameters can have a strong influence on the actual resolution of the acquired image and have consequently a considera-ble impact on the performance of the NN. We propose a known-operator learning approach to perform an end-to-end optimization of MR sequence and neural net-work parameters for SR-TSE. This MR-physics-informed training procedure jointly optimizes the radiofrequency pulse train of a proton density- (PD-) and T2-weighted TSE and a subsequently applied convolutional neural network to predict the corresponding PDw and T2w super-resolution TSE images. The found radiofrequency pulse train designs generate an optimal signal for the NN to perform the SR task. Our method generalizes from the simulation-based optimi-zation to in vivo measurements and the acquired physics-informed SR images show higher correlation with a time-consuming segmented high-resolution TSE sequence compared to a pure network training approach.
Scale-Equivariant Deep Learning for 3D Data
Apr 12, 2023
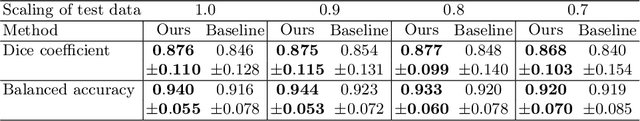
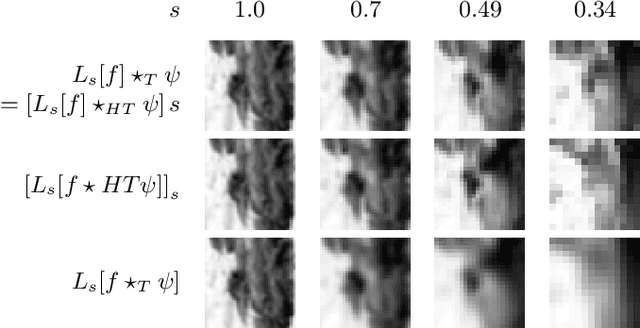
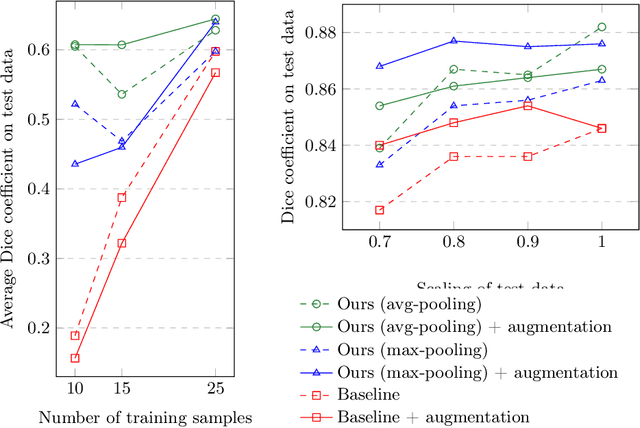
Abstract:The ability of convolutional neural networks (CNNs) to recognize objects regardless of their position in the image is due to the translation-equivariance of the convolutional operation. Group-equivariant CNNs transfer this equivariance to other transformations of the input. Dealing appropriately with objects and object parts of different scale is challenging, and scale can vary for multiple reasons such as the underlying object size or the resolution of the imaging modality. In this paper, we propose a scale-equivariant convolutional network layer for three-dimensional data that guarantees scale-equivariance in 3D CNNs. Scale-equivariance lifts the burden of having to learn each possible scale separately, allowing the neural network to focus on higher-level learning goals, which leads to better results and better data-efficiency. We provide an overview of the theoretical foundations and scientific work on scale-equivariant neural networks in the two-dimensional domain. We then transfer the concepts from 2D to the three-dimensional space and create a scale-equivariant convolutional layer for 3D data. Using the proposed scale-equivariant layer, we create a scale-equivariant U-Net for medical image segmentation and compare it with a non-scale-equivariant baseline method. Our experiments demonstrate the effectiveness of the proposed method in achieving scale-equivariance for 3D medical image analysis. We publish our code at https://github.com/wimmerth/scale-equivariant-3d-convnet for further research and application.
Scene Graph Generation for Better Image Captioning?
Sep 23, 2021
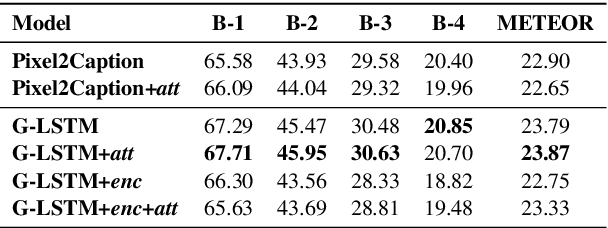
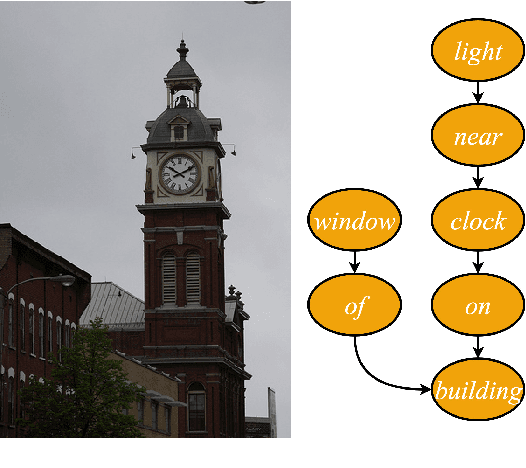
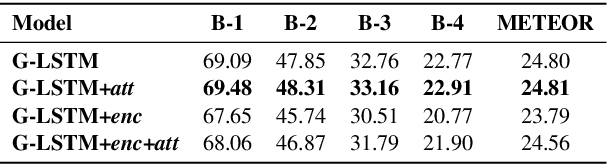
Abstract:We investigate the incorporation of visual relationships into the task of supervised image caption generation by proposing a model that leverages detected objects and auto-generated visual relationships to describe images in natural language. To do so, we first generate a scene graph from raw image pixels by identifying individual objects and visual relationships between them. This scene graph then serves as input to our graph-to-text model, which generates the final caption. In contrast to previous approaches, our model thus explicitly models the detection of objects and visual relationships in the image. For our experiments we construct a new dataset from the intersection of Visual Genome and MS COCO, consisting of images with both a corresponding gold scene graph and human-authored caption. Our results show that our methods outperform existing state-of-the-art end-to-end models that generate image descriptions directly from raw input pixels when compared in terms of the BLEU and METEOR evaluation metrics.
Rotation-Equivariant Deep Learning for Diffusion MRI
Feb 13, 2021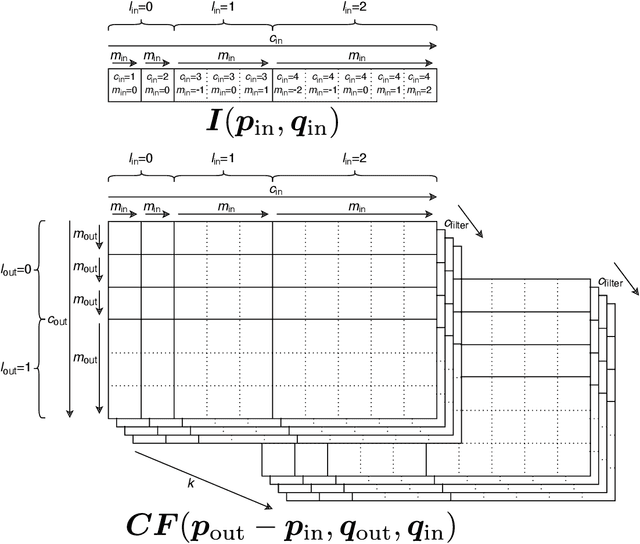
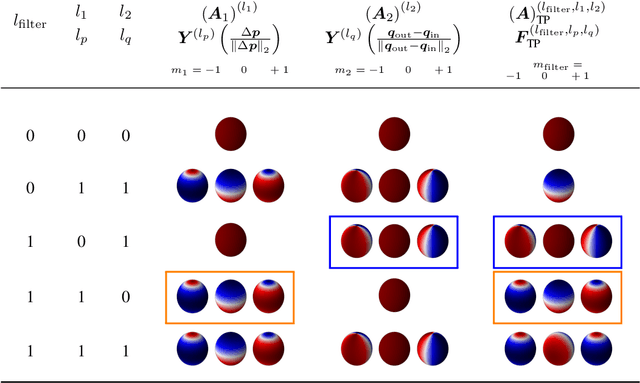
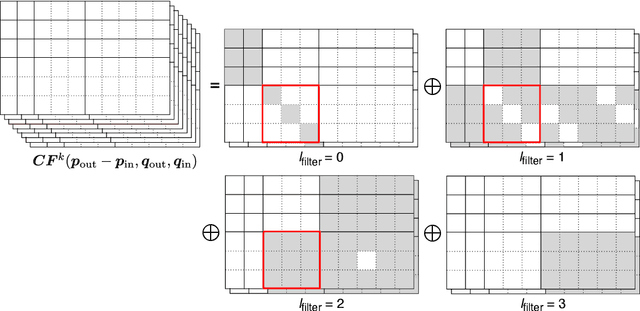
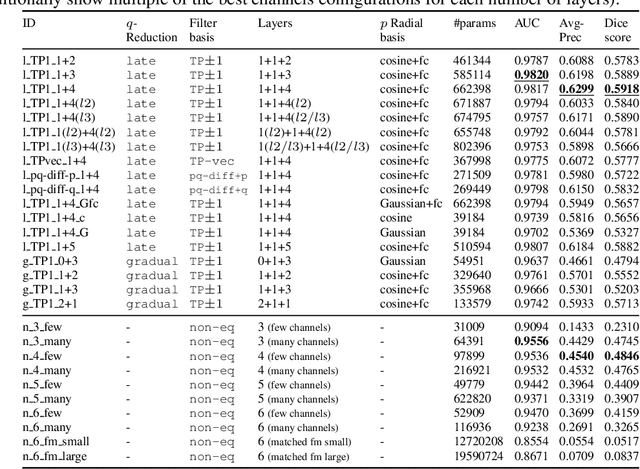
Abstract:Convolutional networks are successful, but they have recently been outperformed by new neural networks that are equivariant under rotations and translations. These new networks work better because they do not struggle with learning each possible orientation of each image feature separately. So far, they have been proposed for 2D and 3D data. Here we generalize them to 6D diffusion MRI data, ensuring joint equivariance under 3D roto-translations in image space and the matching 3D rotations in $q$-space, as dictated by the image formation. Such equivariant deep learning is appropriate for diffusion MRI, because microstructural and macrostructural features such as neural fibers can appear at many different orientations, and because even non-rotation-equivariant deep learning has so far been the best method for many diffusion MRI tasks. We validate our equivariant method on multiple-sclerosis lesion segmentation. Our proposed neural networks yield better results and require fewer scans for training compared to non-rotation-equivariant deep learning. They also inherit all the advantages of deep learning over classical diffusion MRI methods. Our implementation is available at https://github.com/philip-mueller/equivariant-deep-dmri and can be used off the shelf without understanding the mathematical background.
Deep Learning for Virtual Screening: Five Reasons to Use ROC Cost Functions
Jun 25, 2020Abstract:Computer-aided drug discovery is an essential component of modern drug development. Therein, deep learning has become an important tool for rapid screening of billions of molecules in silico for potential hits containing desired chemical features. Despite its importance, substantial challenges persist in training these models, such as severe class imbalance, high decision thresholds, and lack of ground truth labels in some datasets. In this work we argue in favor of directly optimizing the receiver operating characteristic (ROC) in such cases, due to its robustness to class imbalance, its ability to compromise over different decision thresholds, certain freedom to influence the relative weights in this compromise, fidelity to typical benchmarking measures, and equivalence to positive/unlabeled learning. We also propose new training schemes (coherent mini-batch arrangement, and usage of out-of-batch samples) for cost functions based on the ROC, as well as a cost function based on the logAUC metric that facilitates early enrichment (i.e. improves performance at high decision thresholds, as often desired when synthesizing predicted hit compounds). We demonstrate that these approaches outperform standard deep learning approaches on a series of PubChem high-throughput screening datasets that represent realistic and diverse drug discovery campaigns on major drug target families.
Deep Learning for 2D and 3D Rotatable Data: An Overview of Methods
Oct 31, 2019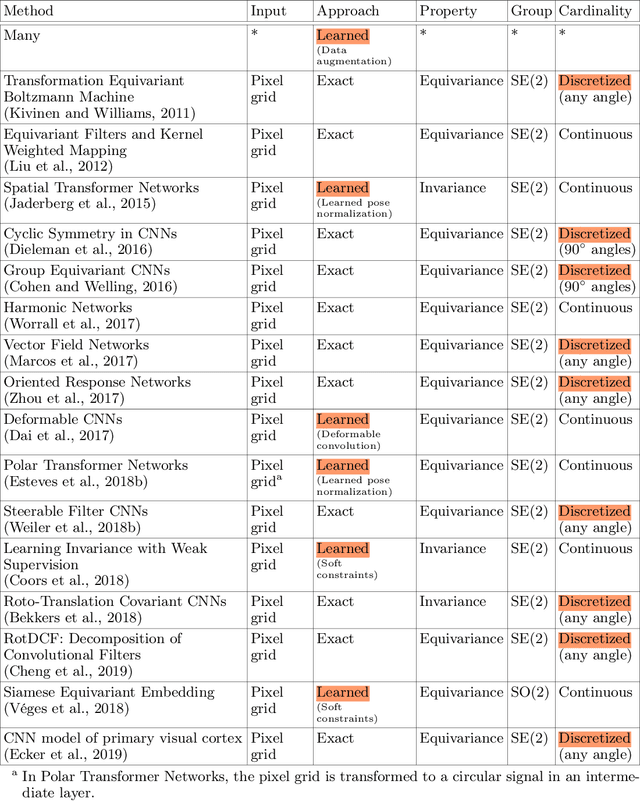
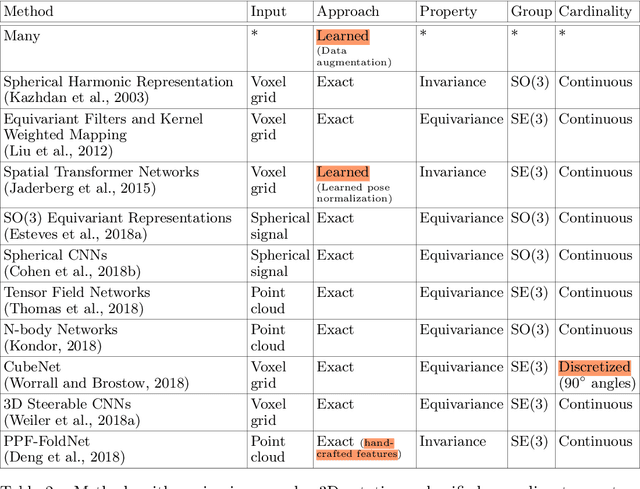
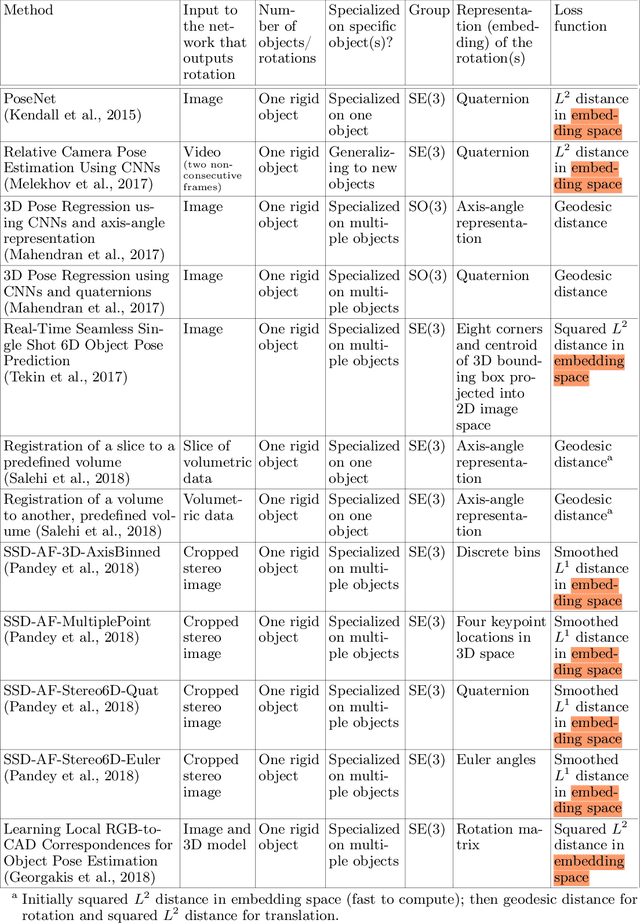
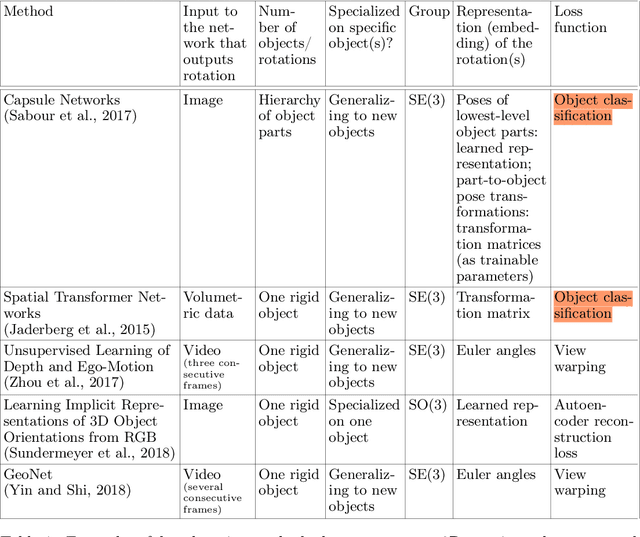
Abstract:One of the reasons for the success of convolutional networks is their equivariance/invariance under translations. However, rotatable data such as molecules, living cells, everyday objects, or galaxies require processing with equivariance/invariance under rotations in cases where the rotation of the coordinate system does not affect the meaning of the data (e.g. object classification). On the other hand, estimation/processing of rotations is necessary in cases where rotations are important (e.g. motion estimation). There has been recent progress in methods and theory in all these regards. Here we provide an overview of existing methods, both for 2D and 3D rotations (and translations), and identify commonalities and links between them, in the hope that our insights will be useful for choosing and perfecting the methods.
Learning to Evolve
May 08, 2019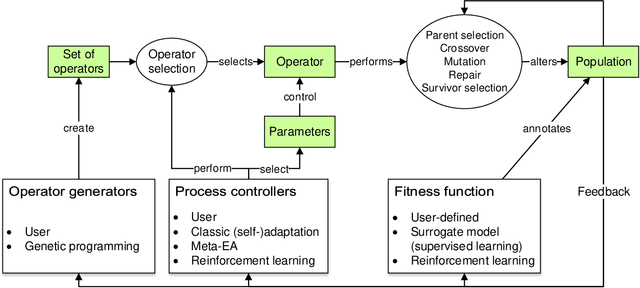
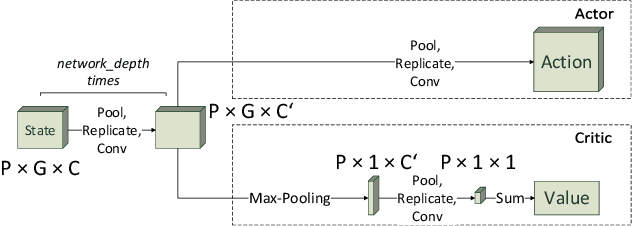
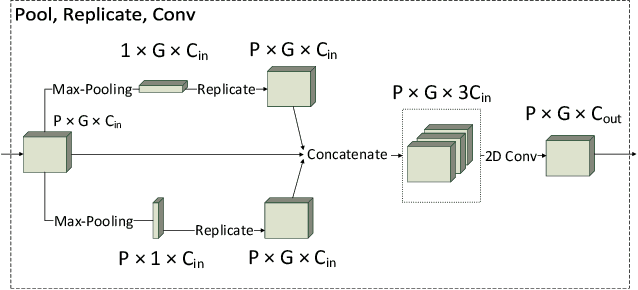
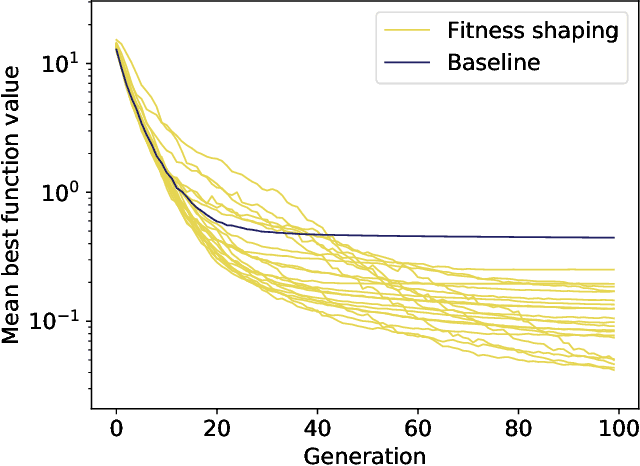
Abstract:Evolution and learning are two of the fundamental mechanisms by which life adapts in order to survive and to transcend limitations. These biological phenomena inspired successful computational methods such as evolutionary algorithms and deep learning. Evolution relies on random mutations and on random genetic recombination. Here we show that learning to evolve, i.e. learning to mutate and recombine better than at random, improves the result of evolution in terms of fitness increase per generation and even in terms of attainable fitness. We use deep reinforcement learning to learn to dynamically adjust the strategy of evolutionary algorithms to varying circumstances. Our methods outperform classical evolutionary algorithms on combinatorial and continuous optimization problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge