Thomas Wimmer
AnyUp: Universal Feature Upsampling
Oct 14, 2025
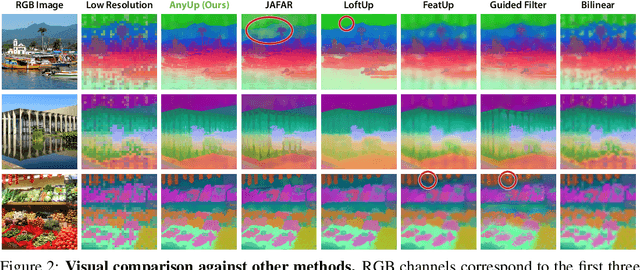

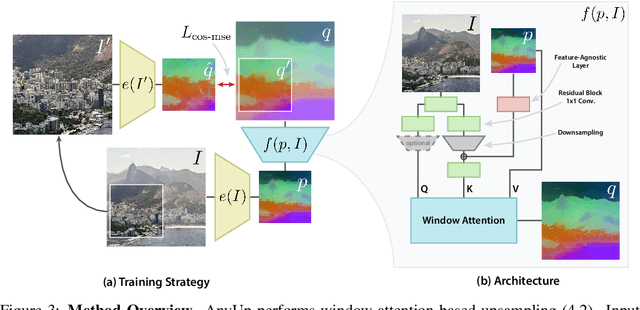
Abstract:We introduce AnyUp, a method for feature upsampling that can be applied to any vision feature at any resolution, without encoder-specific training. Existing learning-based upsamplers for features like DINO or CLIP need to be re-trained for every feature extractor and thus do not generalize to different feature types at inference time. In this work, we propose an inference-time feature-agnostic upsampling architecture to alleviate this limitation and improve upsampling quality. In our experiments, AnyUp sets a new state of the art for upsampled features, generalizes to different feature types, and preserves feature semantics while being efficient and easy to apply to a wide range of downstream tasks.
Do It Yourself: Learning Semantic Correspondence from Pseudo-Labels
Jun 05, 2025Abstract:Finding correspondences between semantically similar points across images and object instances is one of the everlasting challenges in computer vision. While large pre-trained vision models have recently been demonstrated as effective priors for semantic matching, they still suffer from ambiguities for symmetric objects or repeated object parts. We propose to improve semantic correspondence estimation via 3D-aware pseudo-labeling. Specifically, we train an adapter to refine off-the-shelf features using pseudo-labels obtained via 3D-aware chaining, filtering wrong labels through relaxed cyclic consistency, and 3D spherical prototype mapping constraints. While reducing the need for dataset specific annotations compared to prior work, we set a new state-of-the-art on SPair-71k by over 4% absolute gain and by over 7% against methods with similar supervision requirements. The generality of our proposed approach simplifies extension of training to other data sources, which we demonstrate in our experiments.
MEt3R: Measuring Multi-View Consistency in Generated Images
Jan 10, 2025Abstract:We introduce MEt3R, a metric for multi-view consistency in generated images. Large-scale generative models for multi-view image generation are rapidly advancing the field of 3D inference from sparse observations. However, due to the nature of generative modeling, traditional reconstruction metrics are not suitable to measure the quality of generated outputs and metrics that are independent of the sampling procedure are desperately needed. In this work, we specifically address the aspect of consistency between generated multi-view images, which can be evaluated independently of the specific scene. Our approach uses DUSt3R to obtain dense 3D reconstructions from image pairs in a feed-forward manner, which are used to warp image contents from one view into the other. Then, feature maps of these images are compared to obtain a similarity score that is invariant to view-dependent effects. Using MEt3R, we evaluate the consistency of a large set of previous methods for novel view and video generation, including our open, multi-view latent diffusion model.
Gaussians-to-Life: Text-Driven Animation of 3D Gaussian Splatting Scenes
Nov 28, 2024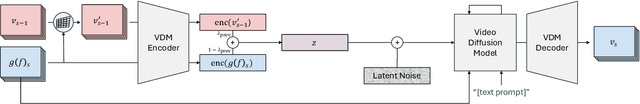
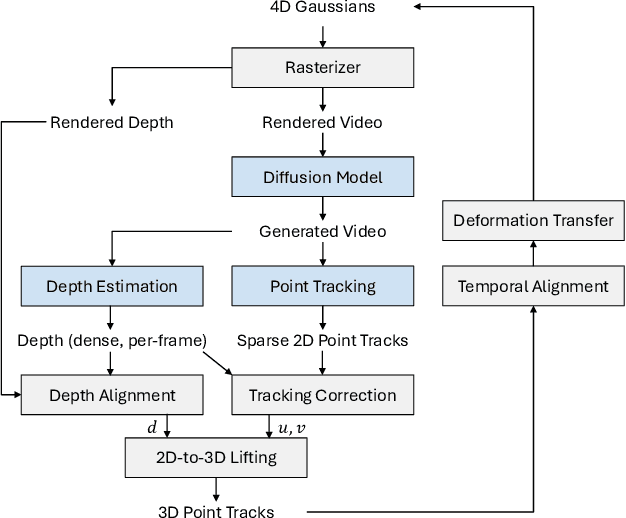
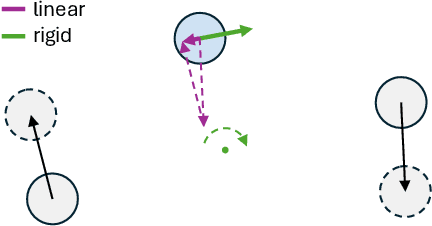
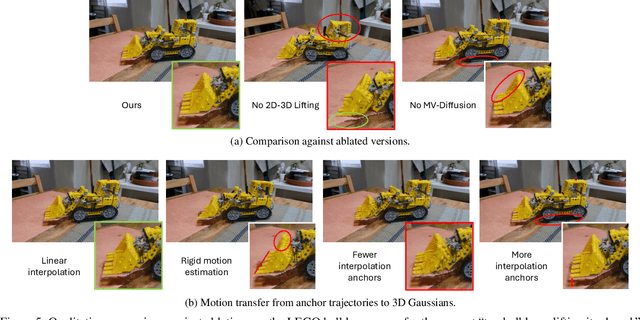
Abstract:State-of-the-art novel view synthesis methods achieve impressive results for multi-view captures of static 3D scenes. However, the reconstructed scenes still lack "liveliness," a key component for creating engaging 3D experiences. Recently, novel video diffusion models generate realistic videos with complex motion and enable animations of 2D images, however they cannot naively be used to animate 3D scenes as they lack multi-view consistency. To breathe life into the static world, we propose Gaussians2Life, a method for animating parts of high-quality 3D scenes in a Gaussian Splatting representation. Our key idea is to leverage powerful video diffusion models as the generative component of our model and to combine these with a robust technique to lift 2D videos into meaningful 3D motion. We find that, in contrast to prior work, this enables realistic animations of complex, pre-existing 3D scenes and further enables the animation of a large variety of object classes, while related work is mostly focused on prior-based character animation, or single 3D objects. Our model enables the creation of consistent, immersive 3D experiences for arbitrary scenes.
Back to 3D: Few-Shot 3D Keypoint Detection with Back-Projected 2D Features
Nov 29, 2023Abstract:With the immense growth of dataset sizes and computing resources in recent years, so-called foundation models have become popular in NLP and vision tasks. In this work, we propose to explore foundation models for the task of keypoint detection on 3D shapes. A unique characteristic of keypoint detection is that it requires semantic and geometric awareness while demanding high localization accuracy. To address this problem, we propose, first, to back-project features from large pre-trained 2D vision models onto 3D shapes and employ them for this task. We show that we obtain robust 3D features that contain rich semantic information and analyze multiple candidate features stemming from different 2D foundation models. Second, we employ a keypoint candidate optimization module which aims to match the average observed distribution of keypoints on the shape and is guided by the back-projected features. The resulting approach achieves a new state of the art for few-shot keypoint detection on the KeyPointNet dataset, almost doubling the performance of the previous best methods.
Language Models for German Text Simplification: Overcoming Parallel Data Scarcity through Style-specific Pre-training
May 22, 2023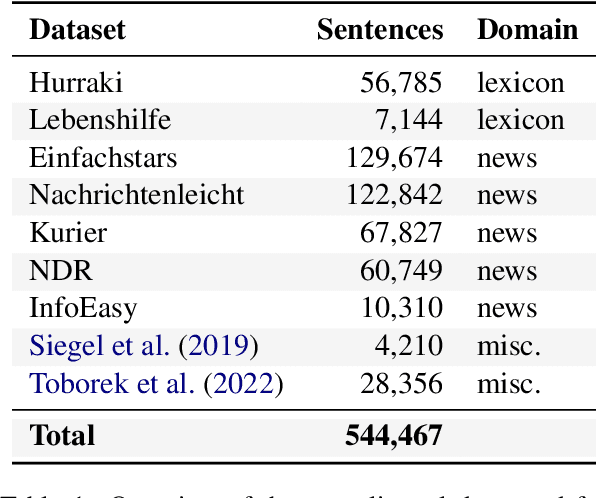
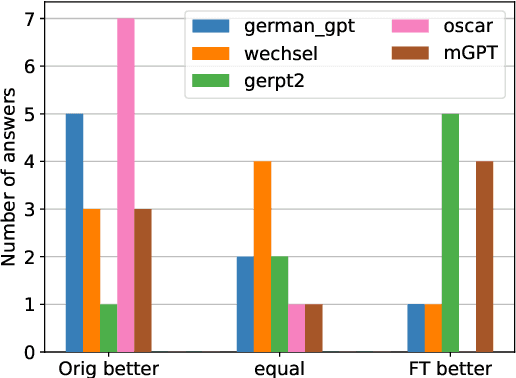
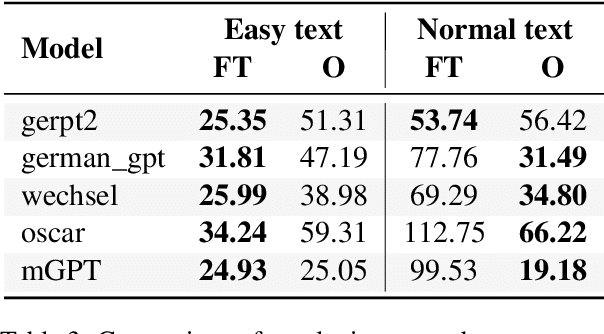
Abstract:Automatic text simplification systems help to reduce textual information barriers on the internet. However, for languages other than English, only few parallel data to train these systems exists. We propose a two-step approach to overcome this data scarcity issue. First, we fine-tuned language models on a corpus of German Easy Language, a specific style of German. Then, we used these models as decoders in a sequence-to-sequence simplification task. We show that the language models adapt to the style characteristics of Easy Language and output more accessible texts. Moreover, with the style-specific pre-training, we reduced the number of trainable parameters in text simplification models. Hence, less parallel data is sufficient for training. Our results indicate that pre-training on unaligned data can reduce the required parallel data while improving the performance on downstream tasks.
Joint MR sequence optimization beats pure neural network approaches for spin-echo MRI super-resolution
May 12, 2023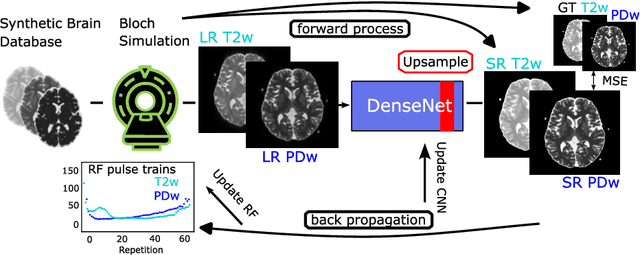
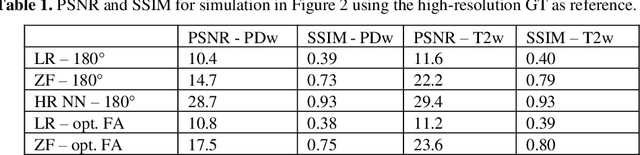
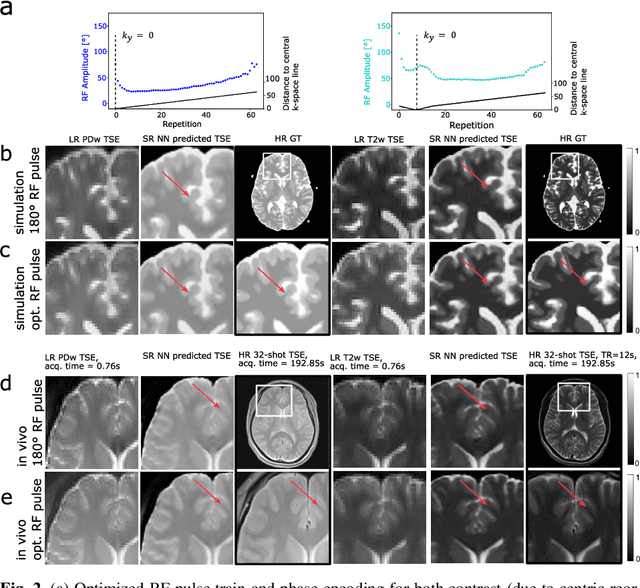
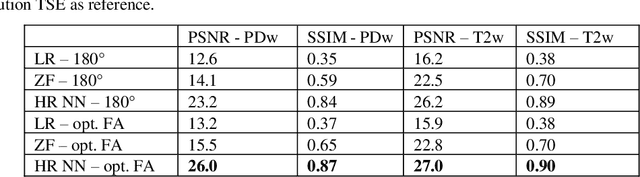
Abstract:Current MRI super-resolution (SR) methods only use existing contrasts acquired from typical clinical sequences as input for the neural network (NN). In turbo spin echo sequences (TSE) the sequence parameters can have a strong influence on the actual resolution of the acquired image and have consequently a considera-ble impact on the performance of the NN. We propose a known-operator learning approach to perform an end-to-end optimization of MR sequence and neural net-work parameters for SR-TSE. This MR-physics-informed training procedure jointly optimizes the radiofrequency pulse train of a proton density- (PD-) and T2-weighted TSE and a subsequently applied convolutional neural network to predict the corresponding PDw and T2w super-resolution TSE images. The found radiofrequency pulse train designs generate an optimal signal for the NN to perform the SR task. Our method generalizes from the simulation-based optimi-zation to in vivo measurements and the acquired physics-informed SR images show higher correlation with a time-consuming segmented high-resolution TSE sequence compared to a pure network training approach.
Scale-Equivariant Deep Learning for 3D Data
Apr 12, 2023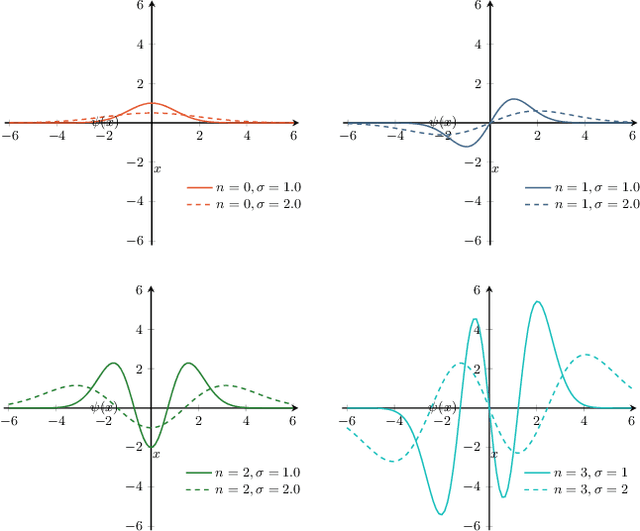
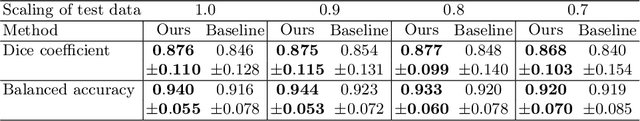
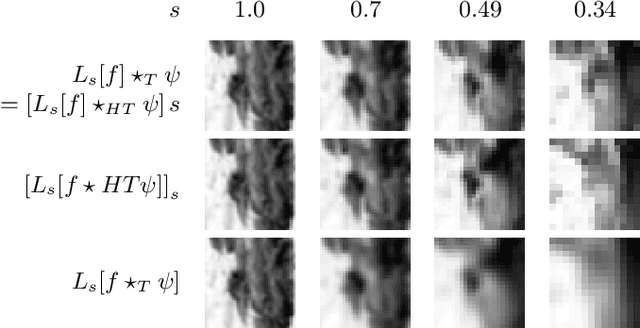
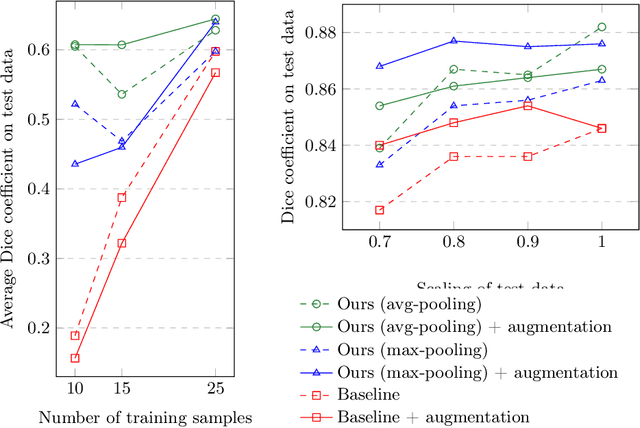
Abstract:The ability of convolutional neural networks (CNNs) to recognize objects regardless of their position in the image is due to the translation-equivariance of the convolutional operation. Group-equivariant CNNs transfer this equivariance to other transformations of the input. Dealing appropriately with objects and object parts of different scale is challenging, and scale can vary for multiple reasons such as the underlying object size or the resolution of the imaging modality. In this paper, we propose a scale-equivariant convolutional network layer for three-dimensional data that guarantees scale-equivariance in 3D CNNs. Scale-equivariance lifts the burden of having to learn each possible scale separately, allowing the neural network to focus on higher-level learning goals, which leads to better results and better data-efficiency. We provide an overview of the theoretical foundations and scientific work on scale-equivariant neural networks in the two-dimensional domain. We then transfer the concepts from 2D to the three-dimensional space and create a scale-equivariant convolutional layer for 3D data. Using the proposed scale-equivariant layer, we create a scale-equivariant U-Net for medical image segmentation and compare it with a non-scale-equivariant baseline method. Our experiments demonstrate the effectiveness of the proposed method in achieving scale-equivariance for 3D medical image analysis. We publish our code at https://github.com/wimmerth/scale-equivariant-3d-convnet for further research and application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge