Or Perlman
Department of Biomedical Engineering, Tel Aviv University, Tel Aviv, Israel, Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel
Multiparameter Uncertainty Mapping in Quantitative Molecular MRI using a Physics-Structured Variational Autoencoder (PS-VAE)
Feb 03, 2026Abstract:Quantitative imaging methods, such as magnetic resonance fingerprinting (MRF), aim to extract interpretable pathology biomarkers by estimating biophysical tissue parameters from signal evolutions. However, the pattern-matching algorithms or neural networks used in such inverse problems often lack principled uncertainty quantification, which limits the trustworthiness and transparency, required for clinical acceptance. Here, we describe a physics-structured variational autoencoder (PS-VAE) designed for rapid extraction of voxelwise multi-parameter posterior distributions. Our approach integrates a differentiable spin physics simulator with self-supervised learning, and provides a full covariance that captures the inter-parameter correlations of the latent biophysical space. The method was validated in a multi-proton pool chemical exchange saturation transfer (CEST) and semisolid magnetization transfer (MT) molecular MRF study, across in-vitro phantoms, tumor-bearing mice, healthy human volunteers, and a subject with glioblastoma. The resulting multi-parametric posteriors are in good agreement with those calculated using a brute-force Bayesian analysis, while providing an orders-of-magnitude acceleration in whole brain quantification. In addition, we demonstrate how monitoring the multi-parameter posterior dynamics across progressively acquired signals provides practical insights for protocol optimization and may facilitate real-time adaptive acquisition.
Multi-Parameter Molecular MRI Quantification using Physics-Informed Self-Supervised Learning
Nov 10, 2024Abstract:Biophysical model fitting plays a key role in obtaining quantitative parameters from physiological signals and images. However, the model complexity for molecular magnetic resonance imaging (MRI) often translates into excessive computation time, which makes clinical use impractical. Here, we present a generic computational approach for solving the parameter extraction inverse problem posed by ordinary differential equation (ODE) modeling coupled with experimental measurement of the system dynamics. This is achieved by formulating a numerical ODE solver to function as a step-wise analytical one, thereby making it compatible with automatic differentiation-based optimization. This enables efficient gradient-based model fitting, and provides a new approach to parameter quantification based on self-supervised learning from a single data observation. The neural-network-based train-by-fit pipeline was used to quantify semisolid magnetization transfer (MT) and chemical exchange saturation transfer (CEST) amide proton exchange parameters in the human brain, in an in-vivo molecular MRI study (n=4). The entire pipeline of the first whole brain quantification was completed in 18.3$\pm$8.3 minutes, which is an order-of-magnitude faster than comparable alternatives. Reusing the single-subject-trained network for inference in new subjects took 1.0$\pm$0.2 s, to provide results in agreement with literature values and scan-specific fit results (Pearson's r>0.98, p<0.0001).
Decoding the human brain tissue response to radiofrequency excitation using a biophysical-model-free deep MRI on a chip framework
Aug 15, 2024



Abstract:Magnetic resonance imaging (MRI) relies on radiofrequency (RF) excitation of proton spin. Clinical diagnosis requires a comprehensive collation of biophysical data via multiple MRI contrasts, acquired using a series of RF sequences that lead to lengthy examinations. Here, we developed a vision transformer-based framework that captures the spatiotemporal magnetic signal evolution and decodes the brain tissue response to RF excitation, constituting an MRI on a chip. Following a per-subject rapid calibration scan (28.2 s), a wide variety of image contrasts including fully quantitative molecular, water relaxation, and magnetic field maps can be generated automatically. The method was validated across healthy subjects and a cancer patient in two different imaging sites, and proved to be 94% faster than alternative protocols. The deep MRI on a chip (DeepMonC) framework may reveal the molecular composition of the human brain tissue in a wide range of pathologies, while offering clinically attractive scan times.
Deep Learning for Accelerated and Robust MRI Reconstruction: a Review
Apr 24, 2024



Abstract:Deep learning (DL) has recently emerged as a pivotal technology for enhancing magnetic resonance imaging (MRI), a critical tool in diagnostic radiology. This review paper provides a comprehensive overview of recent advances in DL for MRI reconstruction. It focuses on DL approaches and architectures designed to improve image quality, accelerate scans, and address data-related challenges. These include end-to-end neural networks, pre-trained networks, generative models, and self-supervised methods. The paper also discusses the role of DL in optimizing acquisition protocols, enhancing robustness against distribution shifts, and tackling subtle bias. Drawing on the extensive literature and practical insights, it outlines current successes, limitations, and future directions for leveraging DL in MRI reconstruction, while emphasizing the potential of DL to significantly impact clinical imaging practices.
Dynamic and Rapid Deep Synthesis of Molecular MRI Signals
May 30, 2023Abstract:Model-driven analysis of biophysical phenomena is gaining increased attention and utility for medical imaging applications. In magnetic resonance imaging (MRI), the availability of well-established models for describing the relations between the nuclear magnetization, tissue properties, and the externally applied magnetic fields has enabled the prediction of image contrast and served as a powerful tool for designing the imaging protocols that are now routinely used in the clinic. Recently, various advanced imaging techniques have relied on these models for image reconstruction, quantitative tissue parameter extraction, and automatic optimization of acquisition protocols. In molecular MRI, however, the increased complexity of the imaging scenario, where the signals from various chemical compounds and multiple proton pools must be accounted for, results in exceedingly long model simulation times, severely hindering the progress of this approach and its dissemination for various clinical applications. Here, we show that a deep-learning-based system can capture the nonlinear relations embedded in the molecular MRI Bloch-McConnell model, enabling a rapid and accurate generation of biologically realistic synthetic data. The applicability of this simulated data for in-silico, in-vitro, and in-vivo imaging applications is then demonstrated for chemical exchange saturation transfer (CEST) and semisolid macromolecule magnetization transfer (MT) analysis and quantification. The proposed approach yielded 78%-99% acceleration in data synthesis time while retaining excellent agreement with the ground truth (Pearson's r$>$0.99, p$<$0.0001, normalized root mean square error $<$3%).
Accelerated and Quantitative 3D Semisolid MT/CEST Imaging using a Generative Adversarial Network (GAN-CEST)
Jul 22, 2022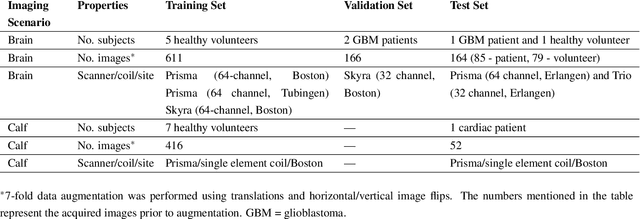
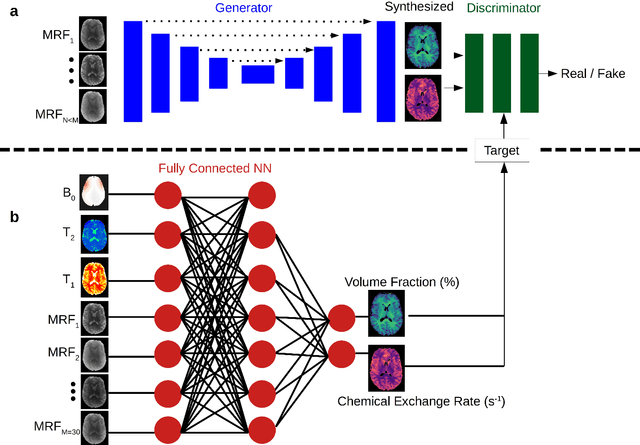
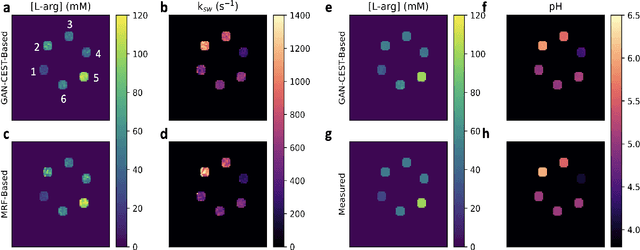
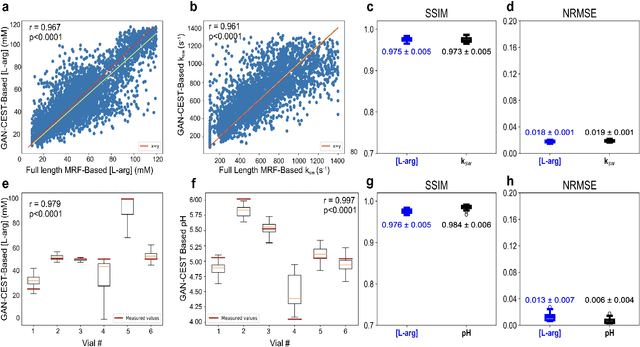
Abstract:Purpose: To substantially shorten the acquisition time required for quantitative 3D chemical exchange saturation transfer (CEST) and semisolid magnetization transfer (MT) imaging and allow for rapid chemical exchange parameter map reconstruction. Methods: Three-dimensional CEST and MT magnetic resonance fingerprinting (MRF) datasets of L-arginine phantoms, whole-brains, and calf muscles from healthy volunteers, cancer patients, and cardiac patients were acquired using 3T clinical scanners at 3 different sites, using 3 different scanner models and coils. A generative adversarial network supervised framework (GAN-CEST) was then designed and trained to learn the mapping from a reduced input data space to the quantitative exchange parameter space, while preserving perceptual and quantitative content. Results: The GAN-CEST 3D acquisition time was 42-52 seconds, 70% shorter than CEST-MRF. The quantitative reconstruction of the entire brain took 0.8 seconds. An excellent agreement was observed between the ground truth and GAN-based L-arginine concentration and pH values (Pearson's r > 0.97, NRMSE < 1.5%). GAN-CEST images from a brain-tumor subject yielded a semi-solid volume fraction and exchange rate NRMSE of 3.8$\pm$1.3% and 4.6$\pm$1.3%, respectively, and SSIM of 96.3$\pm$1.6% and 95.0$\pm$2.4%, respectively. The mapping of the calf-muscle exchange parameters in a cardiac patient, yielded NRMSE < 7% and SSIM > 94% for the semi-solid exchange parameters. In regions with large susceptibility artifacts, GAN-CEST has demonstrated improved performance and reduced noise compared to MRF. Conclusion: GAN-CEST can substantially reduce the acquisition time for quantitative semisolid MT/CEST mapping, while retaining performance even when facing pathologies and scanner models that were not available during training.
Development of a Clinical Chemical Exchange Saturation Transfer MR fingerprinting (CEST-MRF) Pulse Sequence and Reconstruction for Brain Tumor Quantification
Aug 18, 2021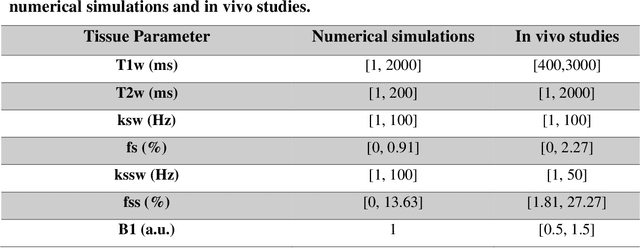



Abstract:Purpose: To develop a clinical chemical exchange saturation transfer magnetic resonance fingerprinting (CEST-MRF) pulse sequence and reconstruction method. Methods: The CEST-MRF pulse sequence was modified to conform to hardware limits on clinical scanners while keeping scan time $\leqslant$ 2 minutes. The measured data was reconstructed using a deep reconstruction network (DRONE) to yield the water relaxation and chemical exchange parameters. The feasibility of the 6 parameter DRONE reconstruction was tested in simulations in a digital brain phantom. A healthy subject was scanned with the CEST-MRF sequence and a conventional MRF sequence for comparison. The reproducibility was assessed via test-retest experiments and the concordance correlation coefficient (CCC) calculated for white matter (WM) and grey matter (GM). The clinical utility of CEST-MRF was demonstrated in a brain metastasis patient in comparison to standard clinical imaging sequences. The tumor was segmented into edema, solid core and necrotic core regions and the CEST-MRF values compared to the contra-lateral side. Results: The 6 parameter DRONE reconstruction of the digital phantom yielded a mean absolute error of $\leqslant$ 6% for all parameters. The CEST-MRF parameters were in good agreement with those from a conventional MRF sequence and previous studies in the literature. The mean CCC for all 6 parameters was 0.79$\pm$0.02 in WM and 0.63$\pm$0.03 in GM. The CEST-MRF values in nearly all tumor regions were significantly different (p=0.001) from each other and the contra-lateral side. Conclusion: The clinical CEST-MRF sequence provides a method for fast simultaneous quantification of multiple tissue parameters in pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge