Elizabeth R. Gerstner
Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA, USA
Accelerated and Quantitative 3D Semisolid MT/CEST Imaging using a Generative Adversarial Network (GAN-CEST)
Jul 22, 2022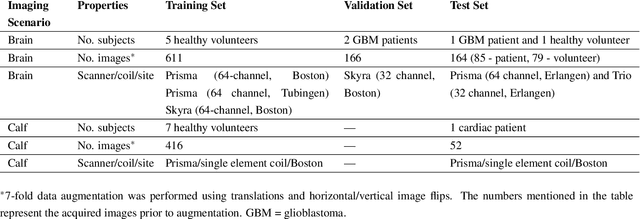
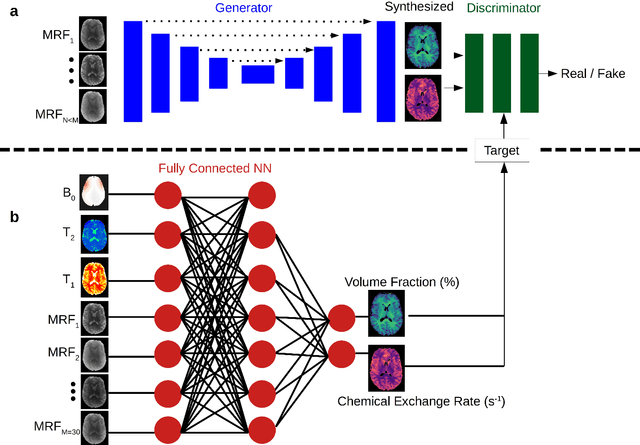
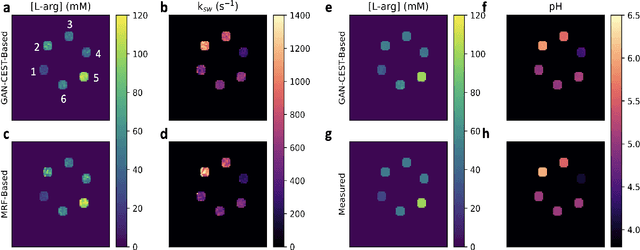
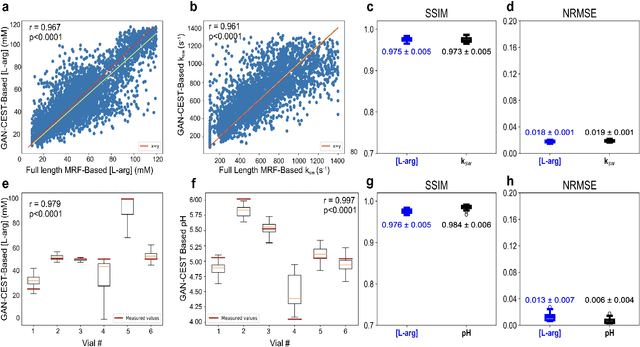
Abstract:Purpose: To substantially shorten the acquisition time required for quantitative 3D chemical exchange saturation transfer (CEST) and semisolid magnetization transfer (MT) imaging and allow for rapid chemical exchange parameter map reconstruction. Methods: Three-dimensional CEST and MT magnetic resonance fingerprinting (MRF) datasets of L-arginine phantoms, whole-brains, and calf muscles from healthy volunteers, cancer patients, and cardiac patients were acquired using 3T clinical scanners at 3 different sites, using 3 different scanner models and coils. A generative adversarial network supervised framework (GAN-CEST) was then designed and trained to learn the mapping from a reduced input data space to the quantitative exchange parameter space, while preserving perceptual and quantitative content. Results: The GAN-CEST 3D acquisition time was 42-52 seconds, 70% shorter than CEST-MRF. The quantitative reconstruction of the entire brain took 0.8 seconds. An excellent agreement was observed between the ground truth and GAN-based L-arginine concentration and pH values (Pearson's r > 0.97, NRMSE < 1.5%). GAN-CEST images from a brain-tumor subject yielded a semi-solid volume fraction and exchange rate NRMSE of 3.8$\pm$1.3% and 4.6$\pm$1.3%, respectively, and SSIM of 96.3$\pm$1.6% and 95.0$\pm$2.4%, respectively. The mapping of the calf-muscle exchange parameters in a cardiac patient, yielded NRMSE < 7% and SSIM > 94% for the semi-solid exchange parameters. In regions with large susceptibility artifacts, GAN-CEST has demonstrated improved performance and reduced noise compared to MRF. Conclusion: GAN-CEST can substantially reduce the acquisition time for quantitative semisolid MT/CEST mapping, while retaining performance even when facing pathologies and scanner models that were not available during training.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021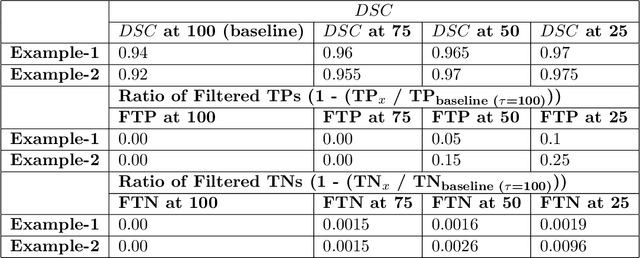
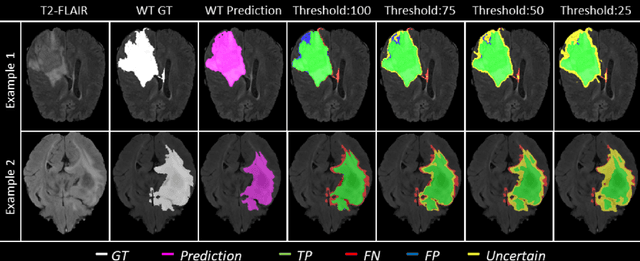
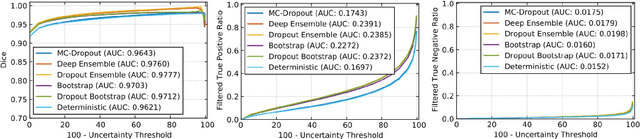
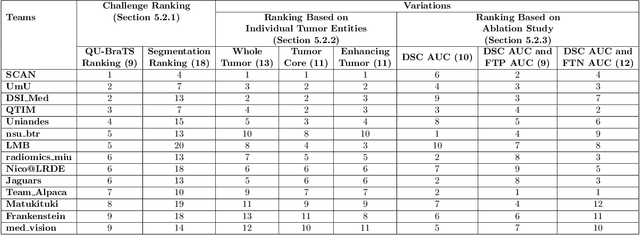
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge