Raphael Meier
A Manually Annotated Image-Caption Dataset for Detecting Children in the Wild
Jun 11, 2025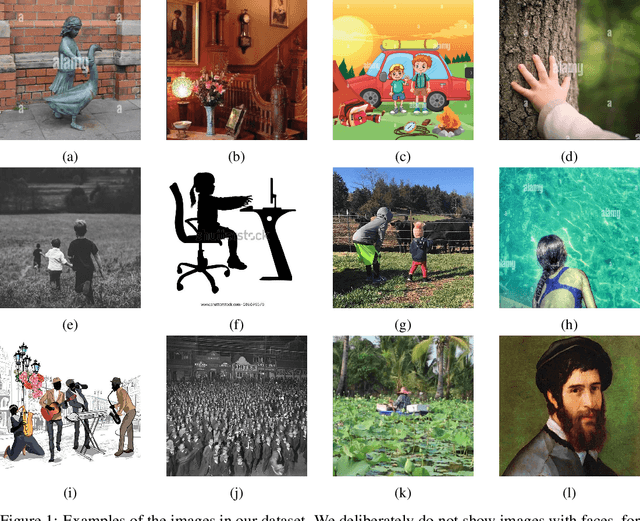
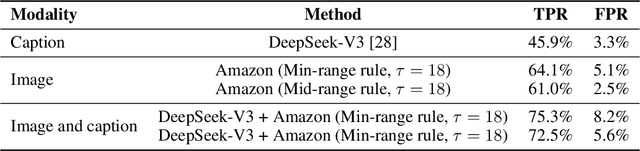

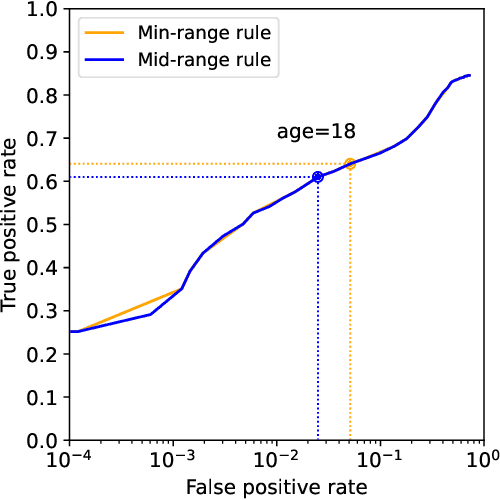
Abstract:Platforms and the law regulate digital content depicting minors (defined as individuals under 18 years of age) differently from other types of content. Given the sheer amount of content that needs to be assessed, machine learning-based automation tools are commonly used to detect content depicting minors. To our knowledge, no dataset or benchmark currently exists for detecting these identification methods in a multi-modal environment. To fill this gap, we release the Image-Caption Children in the Wild Dataset (ICCWD), an image-caption dataset aimed at benchmarking tools that detect depictions of minors. Our dataset is richer than previous child image datasets, containing images of children in a variety of contexts, including fictional depictions and partially visible bodies. ICCWD contains 10,000 image-caption pairs manually labeled to indicate the presence or absence of a child in the image. To demonstrate the possible utility of our dataset, we use it to benchmark three different detectors, including a commercial age estimation system applied to images. Our results suggest that child detection is a challenging task, with the best method achieving a 75.3% true positive rate. We hope the release of our dataset will aid in the design of better minor detection methods in a wide range of scenarios.
Synthetic Photography Detection: A Visual Guidance for Identifying Synthetic Images Created by AI
Aug 12, 2024Abstract:Artificial Intelligence (AI) tools have become incredibly powerful in generating synthetic images. Of particular concern are generated images that resemble photographs as they aspire to represent real world events. Synthetic photographs may be used maliciously by a broad range of threat actors, from scammers to nation-state actors, to deceive, defraud, and mislead people. Mitigating this threat usually involves answering a basic analytic question: Is the photograph real or synthetic? To address this, we have examined the capabilities of recent generative diffusion models and have focused on their flaws: visible artifacts in generated images which reveal their synthetic origin to the trained eye. We categorize these artifacts, provide examples, discuss the challenges in detecting them, suggest practical applications of our work, and outline future research directions.
Sparse vs Contiguous Adversarial Pixel Perturbations in Multimodal Models: An Empirical Analysis
Jul 25, 2024Abstract:Assessing the robustness of multimodal models against adversarial examples is an important aspect for the safety of its users. We craft L0-norm perturbation attacks on the preprocessed input images. We launch them in a black-box setup against four multimodal models and two unimodal DNNs, considering both targeted and untargeted misclassification. Our attacks target less than 0.04% of perturbed image area and integrate different spatial positioning of perturbed pixels: sparse positioning and pixels arranged in different contiguous shapes (row, column, diagonal, and patch). To the best of our knowledge, we are the first to assess the robustness of three state-of-the-art multimodal models (ALIGN, AltCLIP, GroupViT) against different sparse and contiguous pixel distribution perturbations. The obtained results indicate that unimodal DNNs are more robust than multimodal models. Furthermore, models using CNN-based Image Encoder are more vulnerable than models with ViT - for untargeted attacks, we obtain a 99% success rate by perturbing less than 0.02% of the image area.
Synthetic Image Generation in Cyber Influence Operations: An Emergent Threat?
Mar 18, 2024Abstract:The evolution of artificial intelligence (AI) has catalyzed a transformation in digital content generation, with profound implications for cyber influence operations. This report delves into the potential and limitations of generative deep learning models, such as diffusion models, in fabricating convincing synthetic images. We critically assess the accessibility, practicality, and output quality of these tools and their implications in threat scenarios of deception, influence, and subversion. Notably, the report generates content for several hypothetical cyber influence operations to demonstrate the current capabilities and limitations of these AI-driven methods for threat actors. While generative models excel at producing illustrations and non-realistic imagery, creating convincing photo-realistic content remains a significant challenge, limited by computational resources and the necessity for human-guided refinement. Our exploration underscores the delicate balance between technological advancement and its potential for misuse, prompting recommendations for ongoing research, defense mechanisms, multi-disciplinary collaboration, and policy development. These recommendations aim to leverage AI's potential for positive impact while safeguarding against its risks to the integrity of information, especially in the context of cyber influence.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021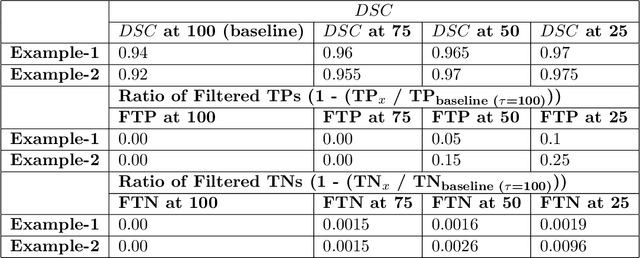
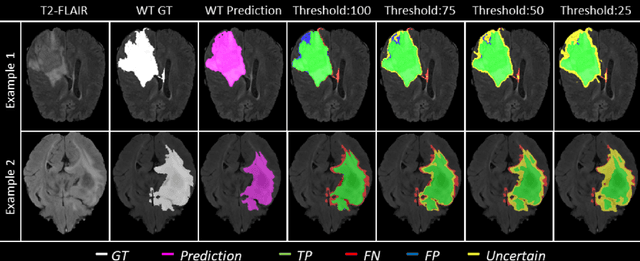
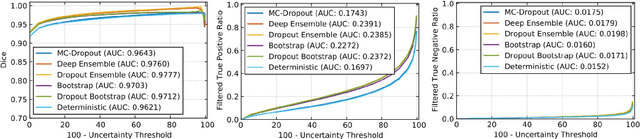
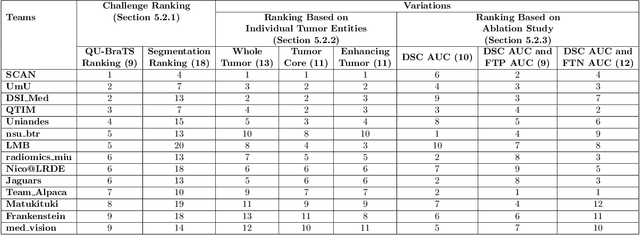
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Combining unsupervised and supervised learning for predicting the final stroke lesion
Jan 02, 2021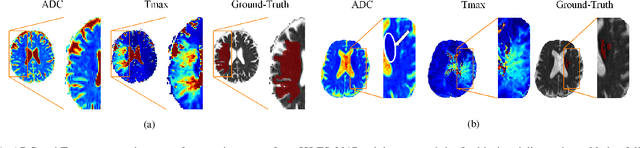
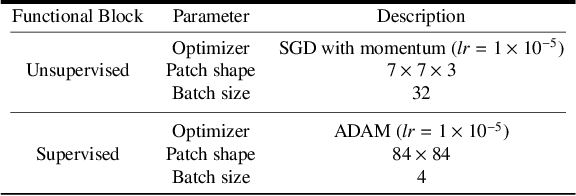
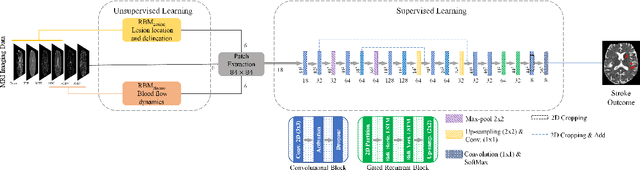
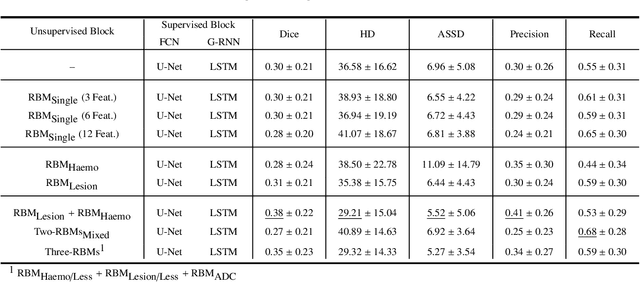
Abstract:Predicting the final ischaemic stroke lesion provides crucial information regarding the volume of salvageable hypoperfused tissue, which helps physicians in the difficult decision-making process of treatment planning and intervention. Treatment selection is influenced by clinical diagnosis, which requires delineating the stroke lesion, as well as characterising cerebral blood flow dynamics using neuroimaging acquisitions. Nonetheless, predicting the final stroke lesion is an intricate task, due to the variability in lesion size, shape, location and the underlying cerebral haemodynamic processes that occur after the ischaemic stroke takes place. Moreover, since elapsed time between stroke and treatment is related to the loss of brain tissue, assessing and predicting the final stroke lesion needs to be performed in a short period of time, which makes the task even more complex. Therefore, there is a need for automatic methods that predict the final stroke lesion and support physicians in the treatment decision process. We propose a fully automatic deep learning method based on unsupervised and supervised learning to predict the final stroke lesion after 90 days. Our aim is to predict the final stroke lesion location and extent, taking into account the underlying cerebral blood flow dynamics that can influence the prediction. To achieve this, we propose a two-branch Restricted Boltzmann Machine, which provides specialized data-driven features from different sets of standard parametric Magnetic Resonance Imaging maps. These data-driven feature maps are then combined with the parametric Magnetic Resonance Imaging maps, and fed to a Convolutional and Recurrent Neural Network architecture. We evaluated our proposal on the publicly available ISLES 2017 testing dataset, reaching a Dice score of 0.38, Hausdorff Distance of 29.21 mm, and Average Symmetric Surface Distance of 5.52 mm.
Uncertainty-driven refinement of tumor-core segmentation using 3D-to-2D networks with label uncertainty
Dec 11, 2020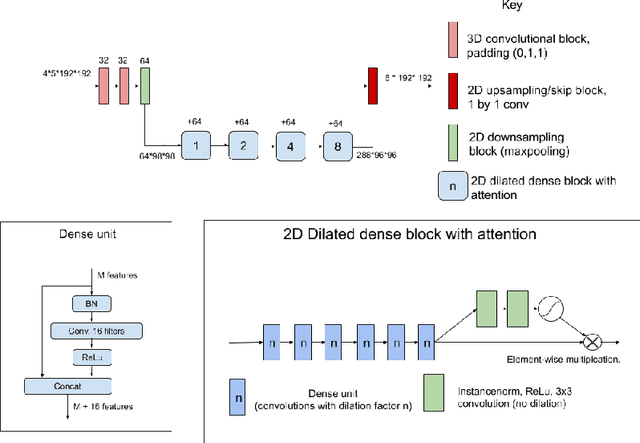
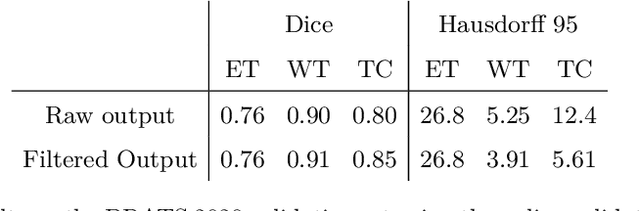
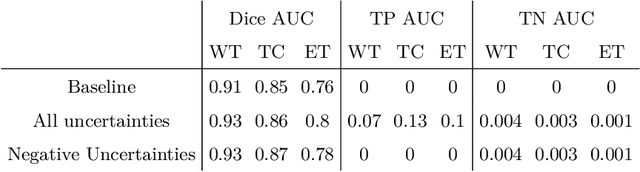
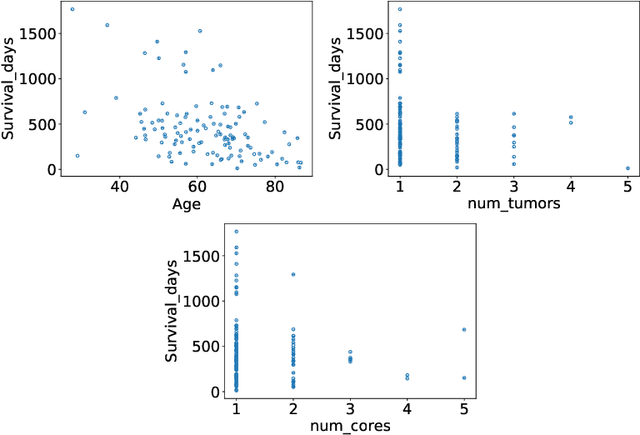
Abstract:The BraTS dataset contains a mixture of high-grade and low-grade gliomas, which have a rather different appearance: previous studies have shown that performance can be improved by separated training on low-grade gliomas (LGGs) and high-grade gliomas (HGGs), but in practice this information is not available at test time to decide which model to use. By contrast with HGGs, LGGs often present no sharp boundary between the tumor core and the surrounding edema, but rather a gradual reduction of tumor-cell density. Utilizing our 3D-to-2D fully convolutional architecture, DeepSCAN, which ranked highly in the 2019 BraTS challenge and was trained using an uncertainty-aware loss, we separate cases into those with a confidently segmented core, and those with a vaguely segmented or missing core. Since by assumption every tumor has a core, we reduce the threshold for classification of core tissue in those cases where the core, as segmented by the classifier, is vaguely defined or missing. We then predict survival of high-grade glioma patients using a fusion of linear regression and random forest classification, based on age, number of distinct tumor components, and number of distinct tumor cores. We present results on the validation dataset of the Multimodal Brain Tumor Segmentation Challenge 2020 (segmentation and uncertainty challenge), and on the testing set, where the method achieved 4th place in Segmentation, 1st place in uncertainty estimation, and 1st place in Survival prediction.
Few-shot brain segmentation from weakly labeled data with deep heteroscedastic multi-task networks
Apr 04, 2019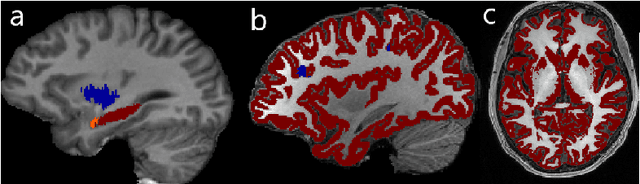
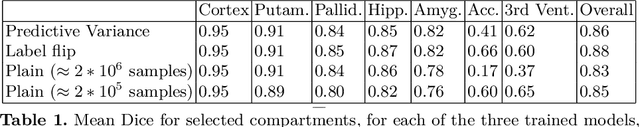
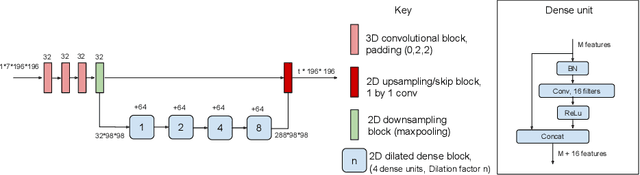
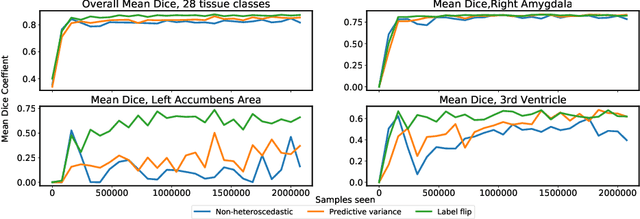
Abstract:In applications of supervised learning applied to medical image segmentation, the need for large amounts of labeled data typically goes unquestioned. In particular, in the case of brain anatomy segmentation, hundreds or thousands of weakly-labeled volumes are often used as training data. In this paper, we first observe that for many brain structures, a small number of training examples, (n=9), weakly labeled using Freesurfer 6.0, plus simple data augmentation, suffice as training data to achieve high performance, achieving an overall mean Dice coefficient of $0.84 \pm 0.12$ compared to Freesurfer over 28 brain structures in T1-weighted images of $\approx 4000$ 9-10 year-olds from the Adolescent Brain Cognitive Development study. We then examine two varieties of heteroscedastic network as a method for improving classification results. An existing proposal by Kendall and Gal, which uses Monte-Carlo inference to learn to predict the variance of each prediction, yields an overall mean Dice of $0.85 \pm 0.14$ and showed statistically significant improvements over 25 brain structures. Meanwhile a novel heteroscedastic network which directly learns the probability that an example has been mislabeled yielded an overall mean Dice of $0.87 \pm 0.11$ and showed statistically significant improvements over all but one of the brain structures considered. The loss function associated to this network can be interpreted as performing a form of learned label smoothing, where labels are only smoothed where they are judged to be uncertain.
Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment
Sep 25, 2018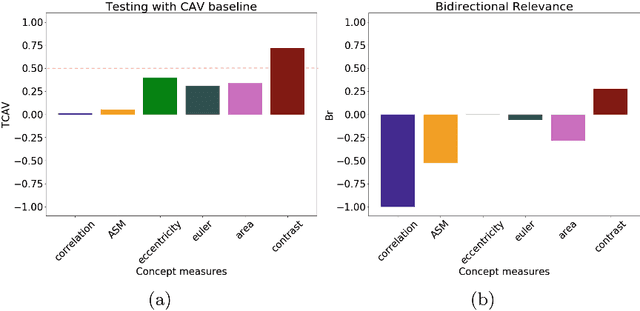
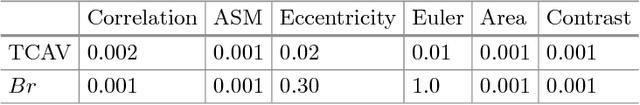
Abstract:Glioblastoma Multiforme is a high grade, very aggressive, brain tumor, with patients having a poor prognosis. Lower grade gliomas are less aggressive, but they can evolve into higher grade tumors over time. Patient management and treatment can vary considerably with tumor grade, ranging from tumor resection followed by a combined radio- and chemotherapy to a "wait and see" approach. Hence, tumor grading is important for adequate treatment planning and monitoring. The gold standard for tumor grading relies on histopathological diagnosis of biopsy specimens. However, this procedure is invasive, time consuming, and prone to sampling error. Given these disadvantages, automatic tumor grading from widely used MRI protocols would be clinically important, as a way to expedite treatment planning and assessment of tumor evolution. In this paper, we propose to use Convolutional Neural Networks for predicting tumor grade directly from imaging data. In this way, we overcome the need for expert annotations of regions of interest. We evaluate two prediction approaches: from the whole brain, and from an automatically defined tumor region. Finally, we employ interpretability methodologies as a quality assurance stage to check if the method is using image regions indicative of tumor grade for classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge