Richard McKinley
Exploring Robustness of Cortical Morphometry in the presence of white matter lesions, using Diffusion Models for Lesion Filling
Mar 26, 2025Abstract:Cortical thickness measurements from magnetic resonance imaging, an important biomarker in many neurodegenerative and neurological disorders, are derived by many tools from an initial voxel-wise tissue segmentation. White matter (WM) hypointensities in T1-weighted imaging, such as those arising from multiple sclerosis or small vessel disease, are known to affect the output of brain segmentation methods and therefore bias cortical thickness measurements. These effects are well-documented among traditional brain segmentation tools but have not been studied extensively in tools based on deep-learning segmentations, which promise to be more robust. In this paper, we explore the potential of deep learning to enhance the accuracy and efficiency of cortical thickness measurement in the presence of WM lesions, using a high-quality lesion filling algorithm leveraging denoising diffusion networks. A pseudo-3D U-Net architecture trained on the OASIS dataset to generate synthetic healthy tissue, conditioned on binary lesion masks derived from the MSSEG dataset, allows realistic removal of white matter lesions in multiple sclerosis patients. By applying morphometry methods to patient images before and after lesion filling, we analysed robustness of global and regional cortical thickness measurements in the presence of white matter lesions. Methods based on a deep learning-based segmentation of the brain (Fastsurfer, DL+DiReCT, ANTsPyNet) exhibited greater robustness than those using classical segmentation methods (Freesurfer, ANTs).
Isometric Transformations for Image Augmentation in Mueller Matrix Polarimetry
Nov 12, 2024



Abstract:Mueller matrix polarimetry captures essential information about polarized light interactions with a sample, presenting unique challenges for data augmentation in deep learning due to its distinct structure. While augmentations are an effective and affordable way to enhance dataset diversity and reduce overfitting, standard transformations like rotations and flips do not preserve the polarization properties in Mueller matrix images. To this end, we introduce a versatile simulation framework that applies physically consistent rotations and flips to Mueller matrices, tailored to maintain polarization fidelity. Our experimental results across multiple datasets reveal that conventional augmentations can lead to misleading results when applied to polarimetric data, underscoring the necessity of our physics-based approach. In our experiments, we first compare our polarization-specific augmentations against real-world captures to validate their physical consistency. We then apply these augmentations in a semantic segmentation task, achieving substantial improvements in model generalization and performance. This study underscores the necessity of physics-informed data augmentation for polarimetric imaging in deep learning (DL), paving the way for broader adoption and more robust applications across diverse research in the field. In particular, our framework unlocks the potential of DL models for polarimetric datasets with limited sample sizes. Our code implementation is available at github.com/hahnec/polar_augment.
Near-Real-Time Mueller Polarimetric Image Processing for Neurosurgical Intervention
Mar 01, 2024Abstract:Wide-field imaging Mueller polarimetry is a revolutionary, label-free, and non-invasive modality for computer-aided intervention: in neurosurgery it aims to provide visual feedback of white matter fibre bundle orientation from derived parameters. Conventionally, robust polarimetric parameters are estimated after averaging multiple measurements of intensity for each pair of probing and detected polarised light. Long multi-shot averaging, however, is not compatible with real-time in-vivo imaging, and the current performance of polarimetric data processing hinders the translation to clinical practice. A learning-based denoising framework is tailored for fast, single-shot, noisy acquisitions of polarimetric intensities. Also, performance-optimised image processing tools are devised for the derivation of clinically relevant parameters. The combination recovers accurate polarimetric parameters from fast acquisitions with near-real-time performance, under the assumption of pseudo-Gaussian polarimetric acquisition noise. The denoising framework is trained, validated, and tested on experimental data comprising tumour-free and diseased human brain samples in different conditions. Accuracy and image quality indices showed significant improvements on testing data for a fast single-pass denoising versus the state-of-the-art and high polarimetric image quality standards. The computational time is reported for the end-to-end processing. The end-to-end image processing achieved real-time performance for a localised field of view. The denoised polarimetric intensities produced visibly clear directional patterns of neuronal fibre tracts in line with reference polarimetric image quality standards; directional disruption was kept in case of neoplastic lesions. The presented advances pave the way towards feasible oncological neurosurgical translations of novel, label free, interventional feedback.
CortexMorph: fast cortical thickness estimation via diffeomorphic registration using VoxelMorph
Jul 21, 2023



Abstract:The thickness of the cortical band is linked to various neurological and psychiatric conditions, and is often estimated through surface-based methods such as Freesurfer in MRI studies. The DiReCT method, which calculates cortical thickness using a diffeomorphic deformation of the gray-white matter interface towards the pial surface, offers an alternative to surface-based methods. Recent studies using a synthetic cortical thickness phantom have demonstrated that the combination of DiReCT and deep-learning-based segmentation is more sensitive to subvoxel cortical thinning than Freesurfer. While anatomical segmentation of a T1-weighted image now takes seconds, existing implementations of DiReCT rely on iterative image registration methods which can take up to an hour per volume. On the other hand, learning-based deformable image registration methods like VoxelMorph have been shown to be faster than classical methods while improving registration accuracy. This paper proposes CortexMorph, a new method that employs unsupervised deep learning to directly regress the deformation field needed for DiReCT. By combining CortexMorph with a deep-learning-based segmentation model, it is possible to estimate region-wise thickness in seconds from a T1-weighted image, while maintaining the ability to detect cortical atrophy. We validate this claim on the OASIS-3 dataset and the synthetic cortical thickness phantom of Rusak et al.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021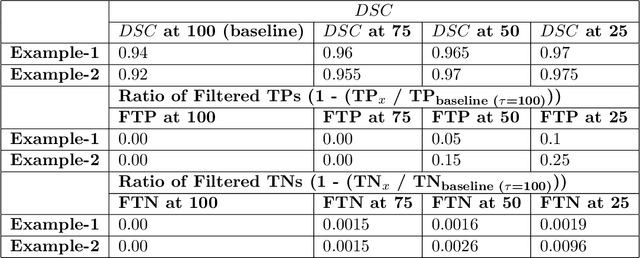
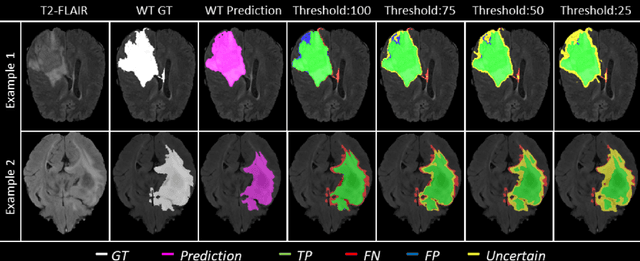
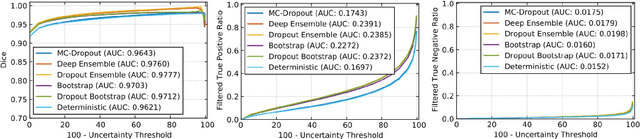
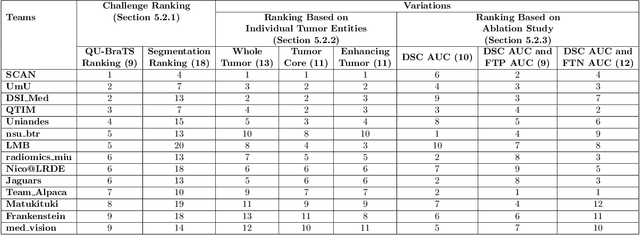
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Are we using appropriate segmentation metrics? Identifying correlates of human expert perception for CNN training beyond rolling the DICE coefficient
Mar 10, 2021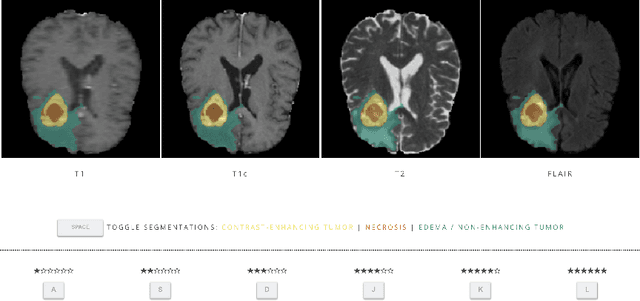

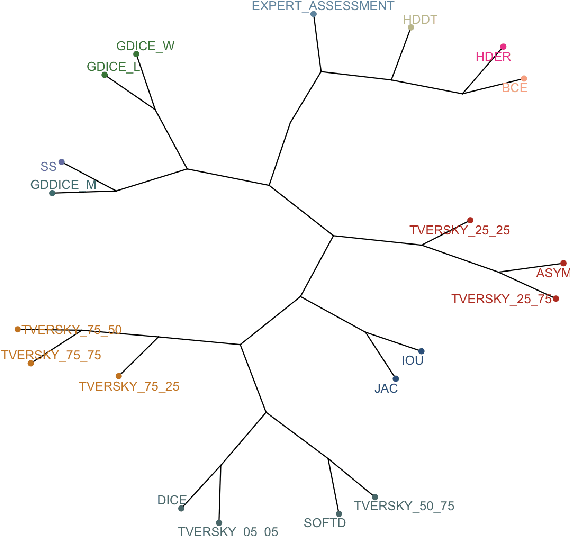
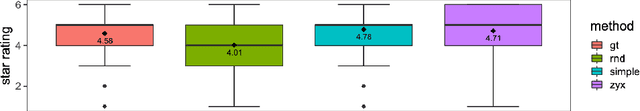
Abstract:In this study, we explore quantitative correlates of qualitative human expert perception. We discover that current quality metrics and loss functions, considered for biomedical image segmentation tasks, correlate moderately with segmentation quality assessment by experts, especially for small yet clinically relevant structures, such as the enhancing tumor in brain glioma. We propose a method employing classical statistics and experimental psychology to create complementary compound loss functions for modern deep learning methods, towards achieving a better fit with human quality assessment. When training a CNN for delineating adult brain tumor in MR images, all four proposed loss candidates outperform the established baselines on the clinically important and hardest to segment enhancing tumor label, while maintaining performance for other label channels.
Uncertainty-driven refinement of tumor-core segmentation using 3D-to-2D networks with label uncertainty
Dec 11, 2020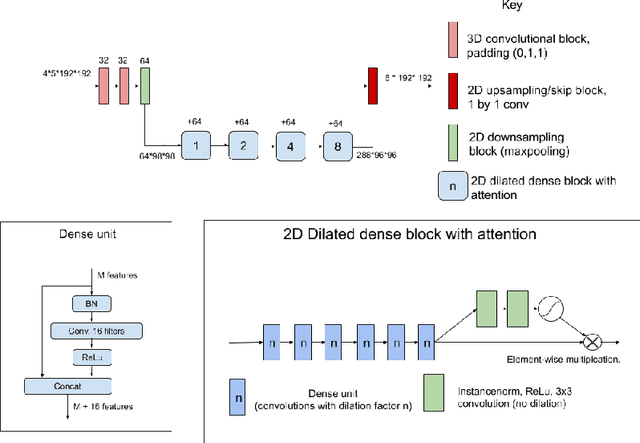
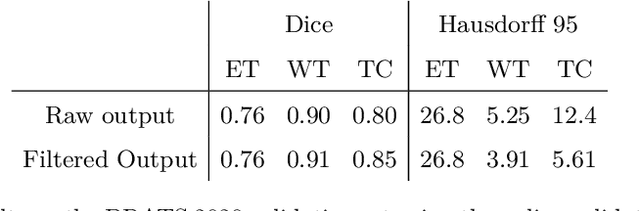
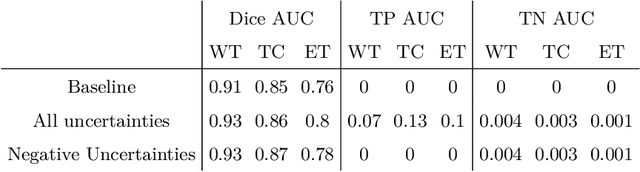
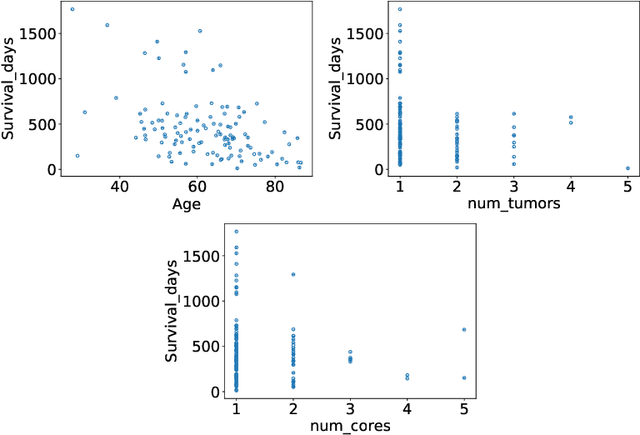
Abstract:The BraTS dataset contains a mixture of high-grade and low-grade gliomas, which have a rather different appearance: previous studies have shown that performance can be improved by separated training on low-grade gliomas (LGGs) and high-grade gliomas (HGGs), but in practice this information is not available at test time to decide which model to use. By contrast with HGGs, LGGs often present no sharp boundary between the tumor core and the surrounding edema, but rather a gradual reduction of tumor-cell density. Utilizing our 3D-to-2D fully convolutional architecture, DeepSCAN, which ranked highly in the 2019 BraTS challenge and was trained using an uncertainty-aware loss, we separate cases into those with a confidently segmented core, and those with a vaguely segmented or missing core. Since by assumption every tumor has a core, we reduce the threshold for classification of core tissue in those cases where the core, as segmented by the classifier, is vaguely defined or missing. We then predict survival of high-grade glioma patients using a fusion of linear regression and random forest classification, based on age, number of distinct tumor components, and number of distinct tumor cores. We present results on the validation dataset of the Multimodal Brain Tumor Segmentation Challenge 2020 (segmentation and uncertainty challenge), and on the testing set, where the method achieved 4th place in Segmentation, 1st place in uncertainty estimation, and 1st place in Survival prediction.
ModelHub.AI: Dissemination Platform for Deep Learning Models
Nov 26, 2019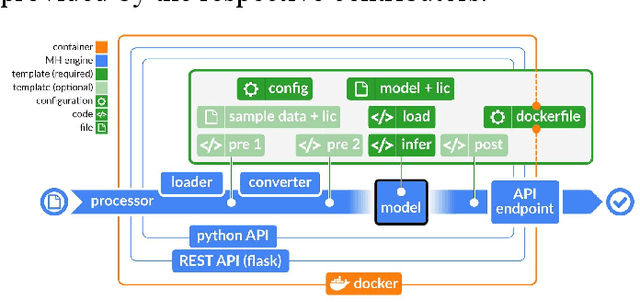
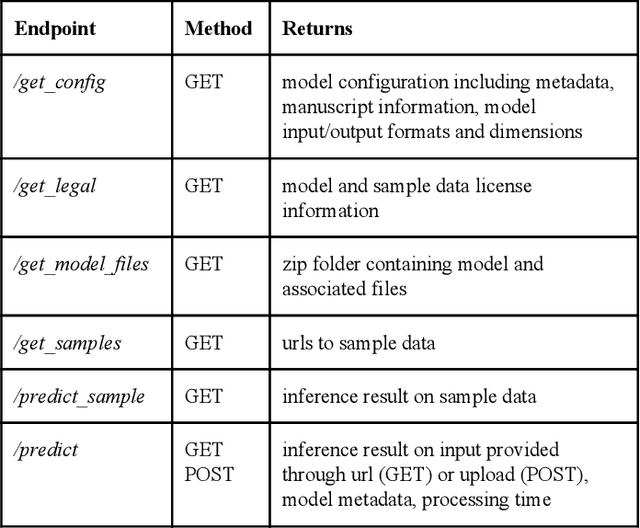
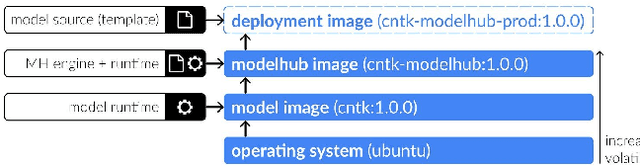
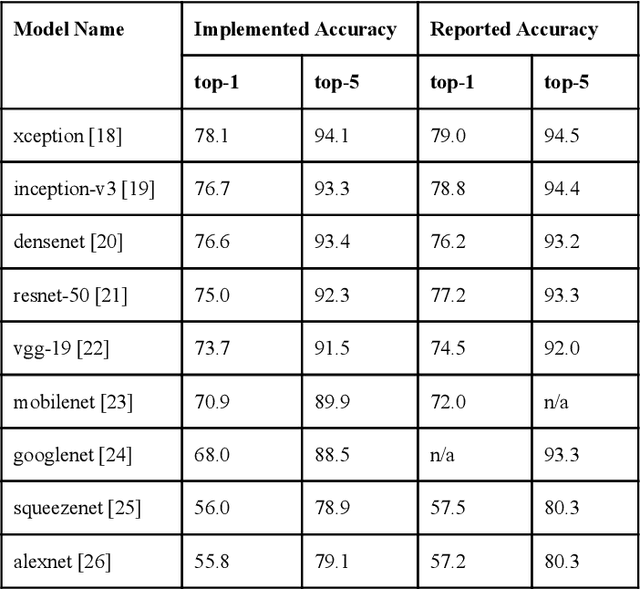
Abstract:Recent advances in artificial intelligence research have led to a profusion of studies that apply deep learning to problems in image analysis and natural language processing among others. Additionally, the availability of open-source computational frameworks has lowered the barriers to implementing state-of-the-art methods across multiple domains. Albeit leading to major performance breakthroughs in some tasks, effective dissemination of deep learning algorithms remains challenging, inhibiting reproducibility and benchmarking studies, impeding further validation, and ultimately hindering their effectiveness in the cumulative scientific progress. In developing a platform for sharing research outputs, we present ModelHub.AI (www.modelhub.ai), a community-driven container-based software engine and platform for the structured dissemination of deep learning models. For contributors, the engine controls data flow throughout the inference cycle, while the contributor-facing standard template exposes model-specific functions including inference, as well as pre- and post-processing. Python and RESTful Application programming interfaces (APIs) enable users to interact with models hosted on ModelHub.AI and allows both researchers and developers to utilize models out-of-the-box. ModelHub.AI is domain-, data-, and framework-agnostic, catering to different workflows and contributors' preferences.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge