Gowtham Krishnan Murugesan
AI generated annotations for Breast, Brain, Liver, Lungs and Prostate cancer collections in National Cancer Institute Imaging Data Commons
Sep 30, 2024



Abstract:AI in Medical Imaging project aims to enhance the National Cancer Institute's (NCI) Image Data Commons (IDC) by developing nnU-Net models and providing AI-assisted segmentations for cancer radiology images. We created high-quality, AI-annotated imaging datasets for 11 IDC collections. These datasets include images from various modalities, such as computed tomography (CT) and magnetic resonance imaging (MRI), covering the lungs, breast, brain, kidneys, prostate, and liver. The nnU-Net models were trained using open-source datasets. A portion of the AI-generated annotations was reviewed and corrected by radiologists. Both the AI and radiologist annotations were encoded in compliance with the the Digital Imaging and Communications in Medicine (DICOM) standard, ensuring seamless integration into the IDC collections. All models, images, and annotations are publicly accessible, facilitating further research and development in cancer imaging. This work supports the advancement of imaging tools and algorithms by providing comprehensive and accurate annotated datasets.
Improving Lesion Segmentation in FDG-18 Whole-Body PET/CT scans using Multilabel approach: AutoPET II challenge
Nov 02, 2023



Abstract:Automatic segmentation of lesions in FDG-18 Whole Body (WB) PET/CT scans using deep learning models is instrumental for determining treatment response, optimizing dosimetry, and advancing theranostic applications in oncology. However, the presence of organs with elevated radiotracer uptake, such as the liver, spleen, brain, and bladder, often leads to challenges, as these regions are often misidentified as lesions by deep learning models. To address this issue, we propose a novel approach of segmenting both organs and lesions, aiming to enhance the performance of automatic lesion segmentation methods. In this study, we assessed the effectiveness of our proposed method using the AutoPET II challenge dataset, which comprises 1014 subjects. We evaluated the impact of inclusion of additional labels and data in the segmentation performance of the model. In addition to the expert-annotated lesion labels, we introduced eight additional labels for organs, including the liver, kidneys, urinary bladder, spleen, lung, brain, heart, and stomach. These labels were integrated into the dataset, and a 3D UNET model was trained within the nnUNet framework. Our results demonstrate that our method achieved the top ranking in the held-out test dataset, underscoring the potential of this approach to significantly improve lesion segmentation accuracy in FDG-18 Whole-Body PET/CT scans, ultimately benefiting cancer patients and advancing clinical practice.
The AIMI Initiative: AI-Generated Annotations for Imaging Data Commons Collections
Oct 23, 2023Abstract:The Image Data Commons (IDC) contains publicly available cancer radiology datasets that could be pertinent to the research and development of advanced imaging tools and algorithms. However, the full extent of its research capabilities is limited by the fact that these datasets have few, if any, annotations associated with them. Through this study with the AI in Medical Imaging (AIMI) initiative a significant contribution, in the form of AI-generated annotations, was made to provide 11 distinct medical imaging collections from the IDC with annotations. These collections included computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) imaging modalities. The main focus of these annotations were in the chest, breast, kidneys, prostate, and liver. Both publicly available and novel AI algorithms were adopted and further developed using open-sourced data coupled with expert annotations to create the AI-generated annotations. A portion of the AI annotations were reviewed and corrected by a radiologist to assess the AI models' performances. Both the AI's and the radiologist's annotations conformed to DICOM standards for seamless integration into the IDC collections as third-party analyses. This study further cements the well-documented notion that expansive publicly accessible datasets, in the field of cancer imaging, coupled with AI will aid in increased accessibility as well as reliability for further research and development.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021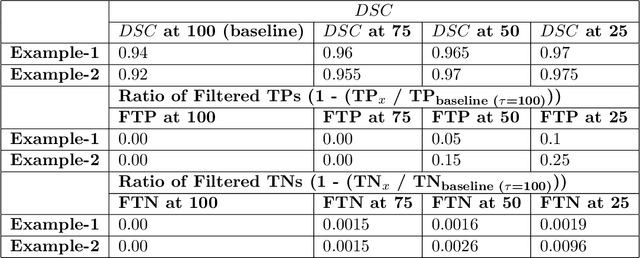
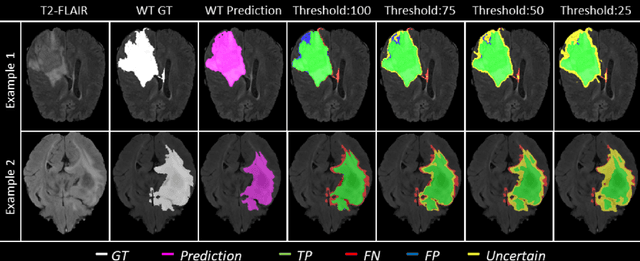
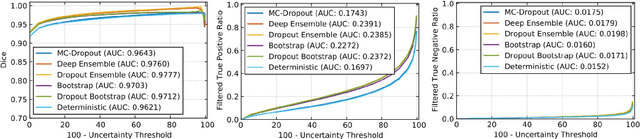
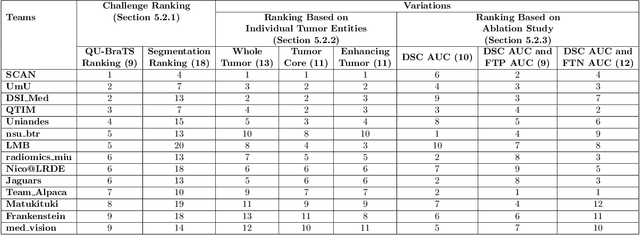
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge