Stephen Moore
AI generated annotations for Breast, Brain, Liver, Lungs and Prostate cancer collections in National Cancer Institute Imaging Data Commons
Sep 30, 2024



Abstract:AI in Medical Imaging project aims to enhance the National Cancer Institute's (NCI) Image Data Commons (IDC) by developing nnU-Net models and providing AI-assisted segmentations for cancer radiology images. We created high-quality, AI-annotated imaging datasets for 11 IDC collections. These datasets include images from various modalities, such as computed tomography (CT) and magnetic resonance imaging (MRI), covering the lungs, breast, brain, kidneys, prostate, and liver. The nnU-Net models were trained using open-source datasets. A portion of the AI-generated annotations was reviewed and corrected by radiologists. Both the AI and radiologist annotations were encoded in compliance with the the Digital Imaging and Communications in Medicine (DICOM) standard, ensuring seamless integration into the IDC collections. All models, images, and annotations are publicly accessible, facilitating further research and development in cancer imaging. This work supports the advancement of imaging tools and algorithms by providing comprehensive and accurate annotated datasets.
Improving Lesion Segmentation in FDG-18 Whole-Body PET/CT scans using Multilabel approach: AutoPET II challenge
Nov 02, 2023



Abstract:Automatic segmentation of lesions in FDG-18 Whole Body (WB) PET/CT scans using deep learning models is instrumental for determining treatment response, optimizing dosimetry, and advancing theranostic applications in oncology. However, the presence of organs with elevated radiotracer uptake, such as the liver, spleen, brain, and bladder, often leads to challenges, as these regions are often misidentified as lesions by deep learning models. To address this issue, we propose a novel approach of segmenting both organs and lesions, aiming to enhance the performance of automatic lesion segmentation methods. In this study, we assessed the effectiveness of our proposed method using the AutoPET II challenge dataset, which comprises 1014 subjects. We evaluated the impact of inclusion of additional labels and data in the segmentation performance of the model. In addition to the expert-annotated lesion labels, we introduced eight additional labels for organs, including the liver, kidneys, urinary bladder, spleen, lung, brain, heart, and stomach. These labels were integrated into the dataset, and a 3D UNET model was trained within the nnUNet framework. Our results demonstrate that our method achieved the top ranking in the held-out test dataset, underscoring the potential of this approach to significantly improve lesion segmentation accuracy in FDG-18 Whole-Body PET/CT scans, ultimately benefiting cancer patients and advancing clinical practice.
English-Twi Parallel Corpus for Machine Translation
Apr 01, 2021


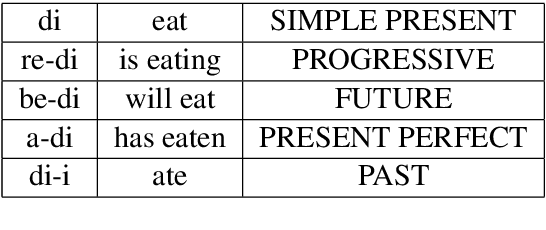
Abstract:We present a parallel machine translation training corpus for English and Akuapem Twi of 25,421 sentence pairs. We used a transformer-based translator to generate initial translations in Akuapem Twi, which were later verified and corrected where necessary by native speakers to eliminate any occurrence of translationese. In addition, 697 higher quality crowd-sourced sentences are provided for use as an evaluation set for downstream Natural Language Processing (NLP) tasks. The typical use case for the larger human-verified dataset is for further training of machine translation models in Akuapem Twi. The higher quality 697 crowd-sourced dataset is recommended as a testing dataset for machine translation of English to Twi and Twi to English models. Furthermore, the Twi part of the crowd-sourced data may also be used for other tasks, such as representation learning, classification, etc. We fine-tune the transformer translation model on the training corpus and report benchmarks on the crowd-sourced test set.
NLP for Ghanaian Languages
Apr 01, 2021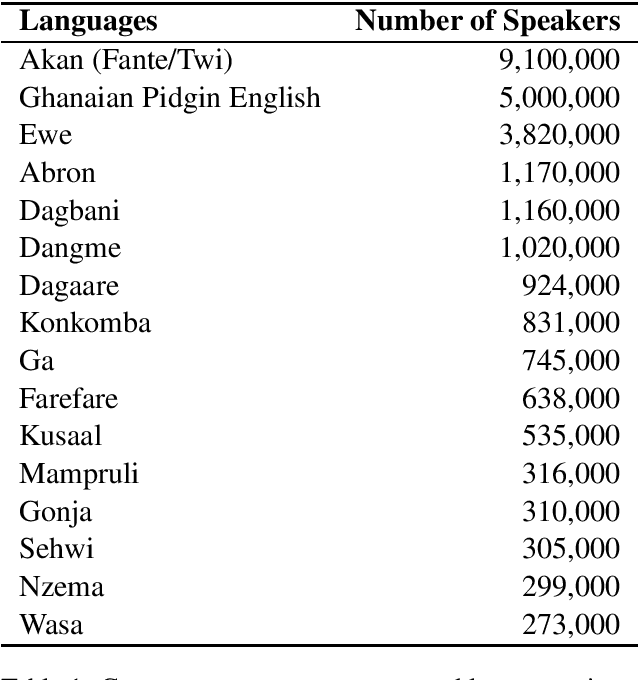
Abstract:NLP Ghana is an open-source non-profit organization aiming to advance the development and adoption of state-of-the-art NLP techniques and digital language tools to Ghanaian languages and problems. In this paper, we first present the motivation and necessity for the efforts of the organization; by introducing some popular Ghanaian languages while presenting the state of NLP in Ghana. We then present the NLP Ghana organization and outline its aims, scope of work, some of the methods employed and contributions made thus far in the NLP community in Ghana.
Contextual Text Embeddings for Twi
Mar 31, 2021
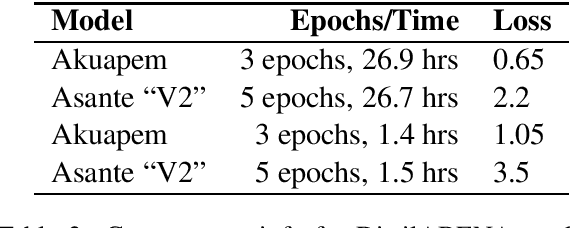


Abstract:Transformer-based language models have been changing the modern Natural Language Processing (NLP) landscape for high-resource languages such as English, Chinese, Russian, etc. However, this technology does not yet exist for any Ghanaian language. In this paper, we introduce the first of such models for Twi or Akan, the most widely spoken Ghanaian language. The specific contribution of this research work is the development of several pretrained transformer language models for the Akuapem and Asante dialects of Twi, paving the way for advances in application areas such as Named Entity Recognition (NER), Neural Machine Translation (NMT), Sentiment Analysis (SA) and Part-of-Speech (POS) tagging. Specifically, we introduce four different flavours of ABENA -- A BERT model Now in Akan that is fine-tuned on a set of Akan corpora, and BAKO - BERT with Akan Knowledge only, which is trained from scratch. We open-source the model through the Hugging Face model hub and demonstrate its use via a simple sentiment classification example.
Deep Semantic Instance Segmentation of Tree-like Structures Using Synthetic Data
Nov 08, 2018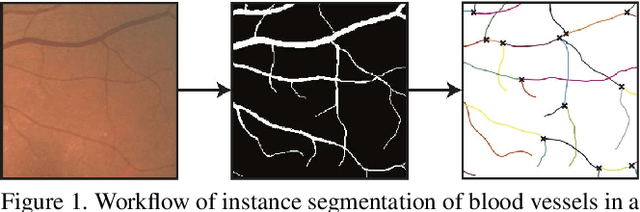
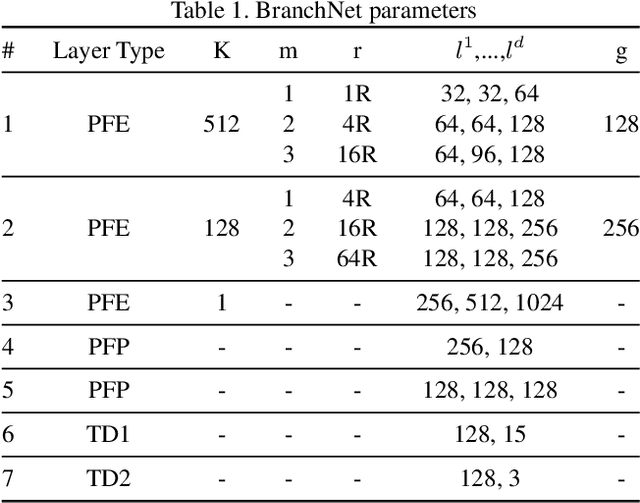
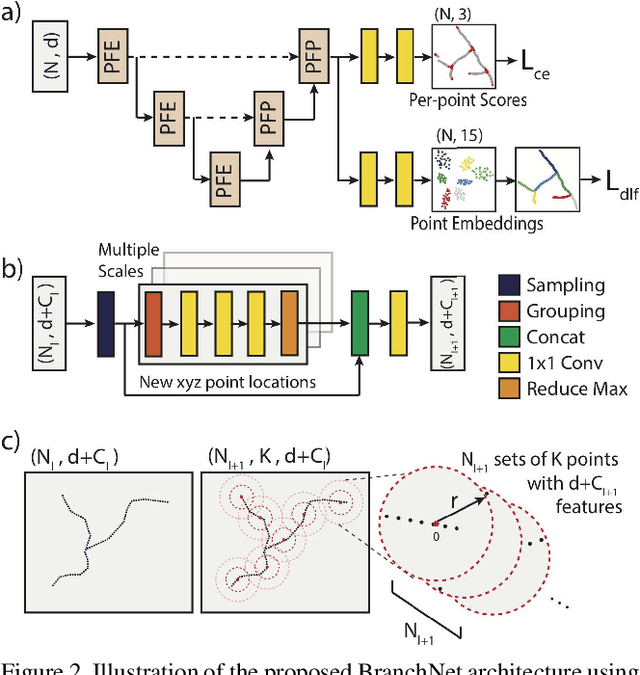
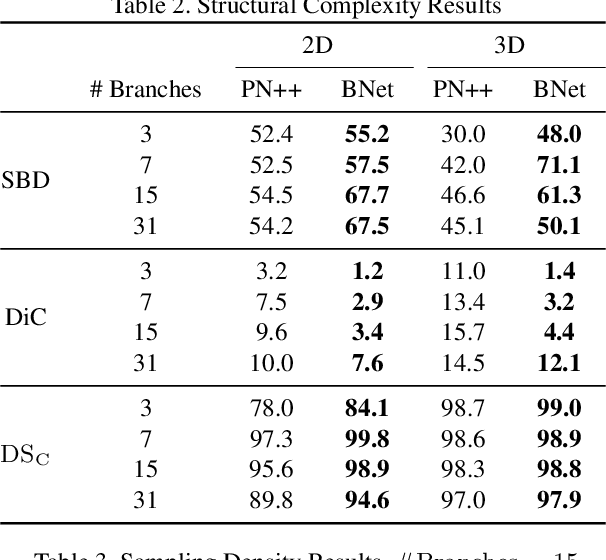
Abstract:Tree-like structures, such as blood vessels, often express complexity at very fine scales, requiring high-resolution grids to adequately describe their shape. Such sparse morphology can alternately be represented by locations of centreline points, but learning from this type of data with deep learning is challenging due to it being unordered, and permutation invariant. In this work, we propose a deep neural network that directly consumes unordered points along the centreline of a branching structure, to identify the topology of the represented structure in a single-shot. Key to our approach is the use of a novel multi-task loss function, enabling instance segmentation of arbitrarily complex branching structures. We train the network solely using synthetically generated data, utilizing domain randomization to facilitate the transfer to real 2D and 3D data. Results show that our network can reliably extract meaningful information about branch locations, bifurcations and endpoints, and sets a new benchmark for semantic instance segmentation in branching structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge