Brandon Edwards
Privacy-Enhancing Collaborative Information Sharing through Federated Learning -- A Case of the Insurance Industry
Feb 22, 2024



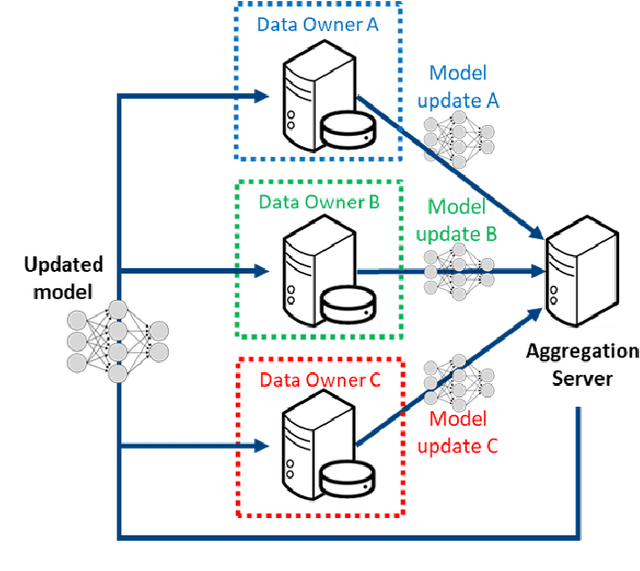
Abstract:The report demonstrates the benefits (in terms of improved claims loss modeling) of harnessing the value of Federated Learning (FL) to learn a single model across multiple insurance industry datasets without requiring the datasets themselves to be shared from one company to another. The application of FL addresses two of the most pressing concerns: limited data volume and data variety, which are caused by privacy concerns, the rarity of claim events, the lack of informative rating factors, etc.. During each round of FL, collaborators compute improvements on the model using their local private data, and these insights are combined to update a global model. Such aggregation of insights allows for an increase to the effectiveness in forecasting claims losses compared to models individually trained at each collaborator. Critically, this approach enables machine learning collaboration without the need for raw data to leave the compute infrastructure of each respective data owner. Additionally, the open-source framework, OpenFL, that is used in our experiments is designed so that it can be run using confidential computing as well as with additional algorithmic protections against leakage of information via the shared model updates. In such a way, FL is implemented as a privacy-enhancing collaborative learning technique that addresses the challenges posed by the sensitivity and privacy of data in traditional machine learning solutions. This paper's application of FL can also be expanded to other areas including fraud detection, catastrophe modeling, etc., that have a similar need to incorporate data privacy into machine learning collaborations. Our framework and empirical results provide a foundation for future collaborations among insurers, regulators, academic researchers, and InsurTech experts.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
The Federated Tumor Segmentation (FeTS) Challenge
May 14, 2021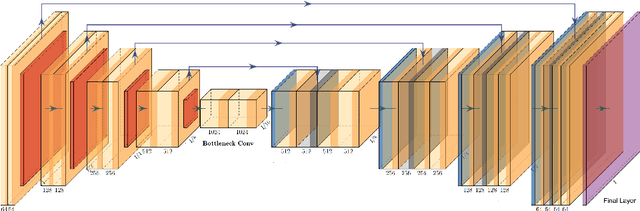
Abstract:This manuscript describes the first challenge on Federated Learning, namely the Federated Tumor Segmentation (FeTS) challenge 2021. International challenges have become the standard for validation of biomedical image analysis methods. However, the actual performance of participating (even the winning) algorithms on "real-world" clinical data often remains unclear, as the data included in challenges are usually acquired in very controlled settings at few institutions. The seemingly obvious solution of just collecting increasingly more data from more institutions in such challenges does not scale well due to privacy and ownership hurdles. Towards alleviating these concerns, we are proposing the FeTS challenge 2021 to cater towards both the development and the evaluation of models for the segmentation of intrinsically heterogeneous (in appearance, shape, and histology) brain tumors, namely gliomas. Specifically, the FeTS 2021 challenge uses clinically acquired, multi-institutional magnetic resonance imaging (MRI) scans from the BraTS 2020 challenge, as well as from various remote independent institutions included in the collaborative network of a real-world federation (https://www.fets.ai/). The goals of the FeTS challenge are directly represented by the two included tasks: 1) the identification of the optimal weight aggregation approach towards the training of a consensus model that has gained knowledge via federated learning from multiple geographically distinct institutions, while their data are always retained within each institution, and 2) the federated evaluation of the generalizability of brain tumor segmentation models "in the wild", i.e. on data from institutional distributions that were not part of the training datasets.
OpenFL: An open-source framework for Federated Learning
May 13, 2021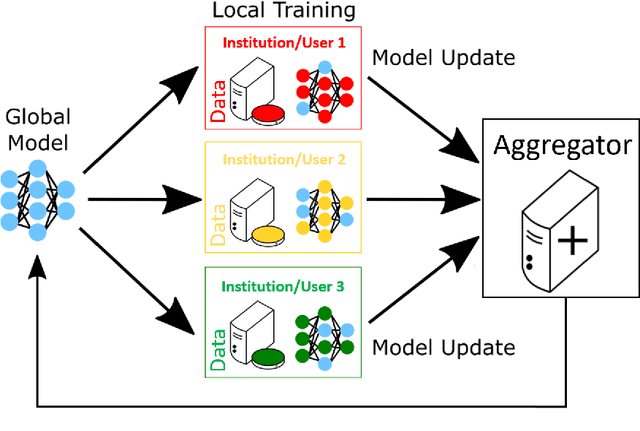
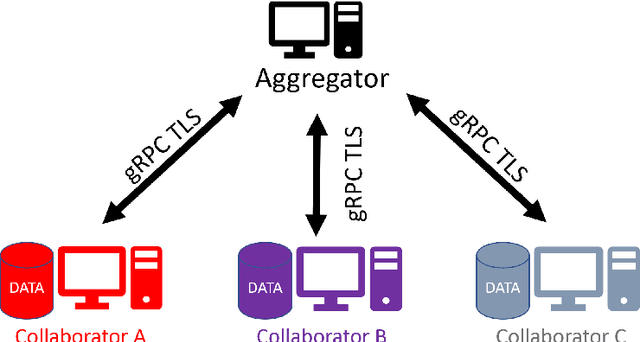
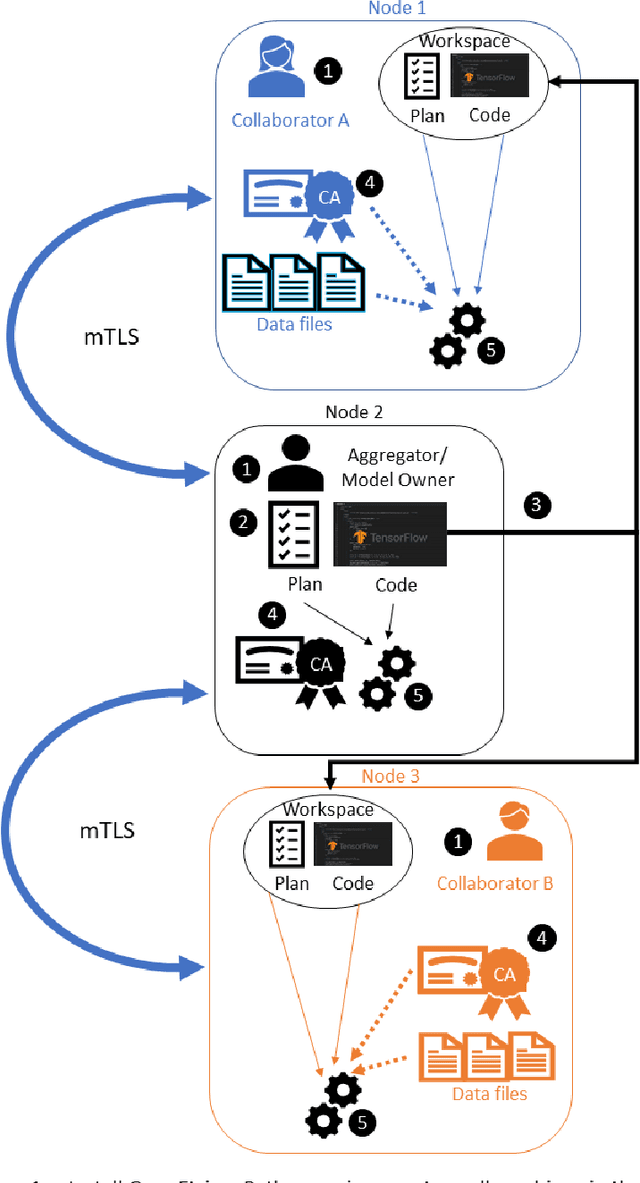
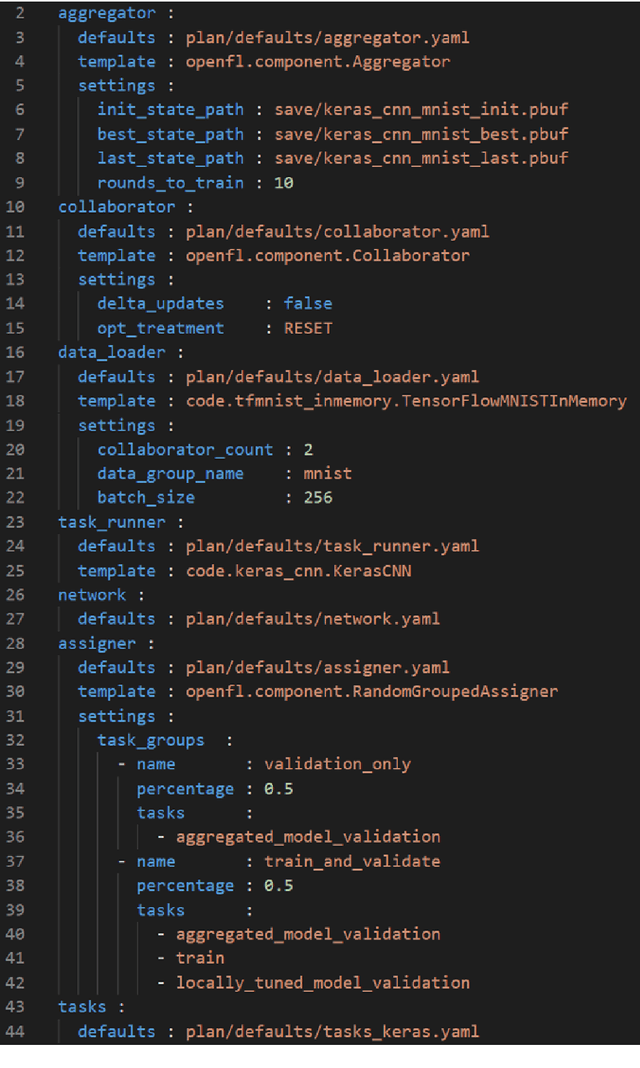
Abstract:Federated learning (FL) is a computational paradigm that enables organizations to collaborate on machine learning (ML) projects without sharing sensitive data, such as, patient records, financial data, or classified secrets. Open Federated Learning (OpenFL https://github.com/intel/openfl) is an open-source framework for training ML algorithms using the data-private collaborative learning paradigm of FL. OpenFL works with training pipelines built with both TensorFlow and PyTorch, and can be easily extended to other ML and deep learning frameworks. Here, we summarize the motivation and development characteristics of OpenFL, with the intention of facilitating its application to existing ML model training in a production environment. Finally, we describe the first use of the OpenFL framework to train consensus ML models in a consortium of international healthcare organizations, as well as how it facilitates the first computational competition on FL.
GaNDLF: A Generally Nuanced Deep Learning Framework for Scalable End-to-End Clinical Workflows in Medical Imaging
Feb 26, 2021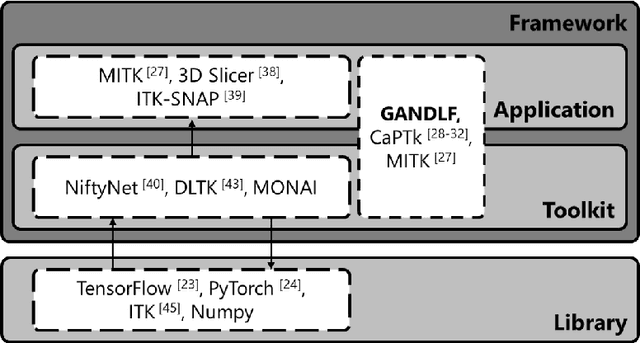
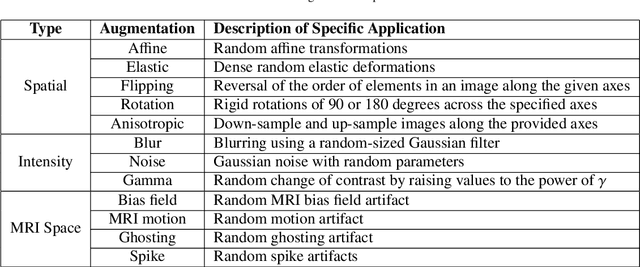
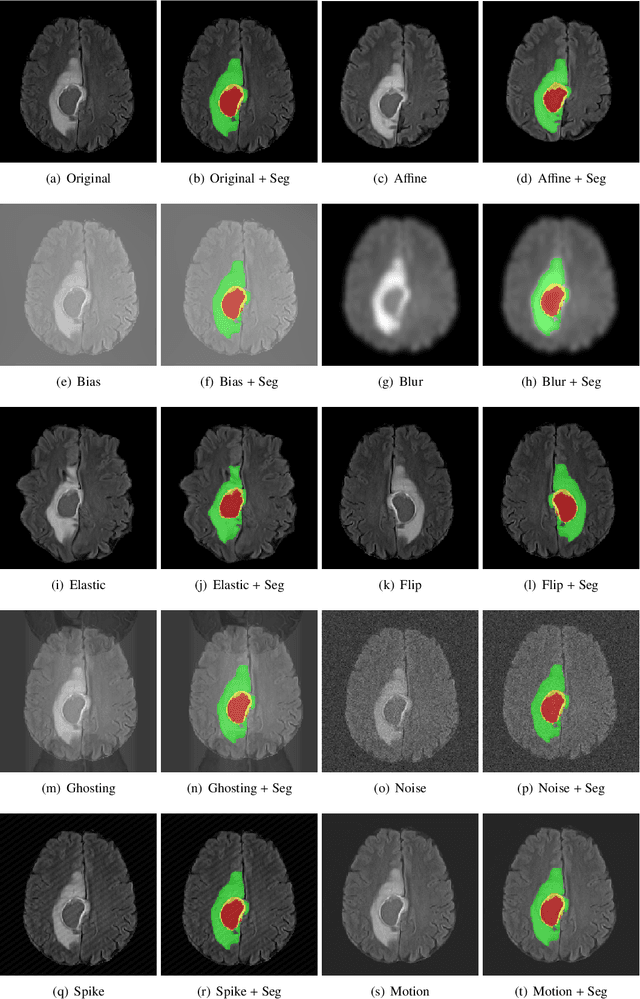
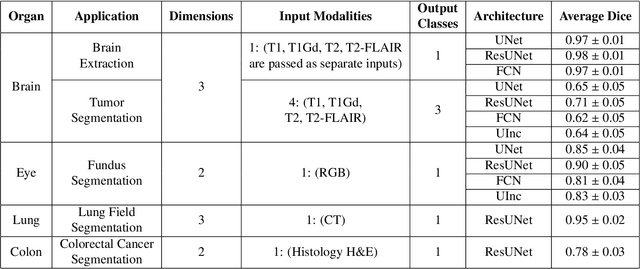
Abstract:Deep Learning (DL) has greatly highlighted the potential impact of optimized machine learning in both the scientific and clinical communities. The advent of open-source DL libraries from major industrial entities, such as TensorFlow (Google), PyTorch (Facebook), and MXNet (Apache), further contributes to DL promises on the democratization of computational analytics. However, increased technical and specialized background is required to develop DL algorithms, and the variability of implementation details hinders their reproducibility. Towards lowering the barrier and making the mechanism of DL development, training, and inference more stable, reproducible, and scalable, without requiring an extensive technical background, this manuscript proposes the \textbf{G}ener\textbf{a}lly \textbf{N}uanced \textbf{D}eep \textbf{L}earning \textbf{F}ramework (GaNDLF). With built-in support for $k$-fold cross-validation, data augmentation, multiple modalities and output classes, and multi-GPU training, as well as the ability to work with both radiographic and histologic imaging, GaNDLF aims to provide an end-to-end solution for all DL-related tasks, to tackle problems in medical imaging and provide a robust application framework for deployment in clinical workflows.
Toward Few-step Adversarial Training from a Frequency Perspective
Oct 13, 2020



Abstract:We investigate adversarial-sample generation methods from a frequency domain perspective and extend standard $l_{\infty}$ Projected Gradient Descent (PGD) to the frequency domain. The resulting method, which we call Spectral Projected Gradient Descent (SPGD), has better success rate compared to PGD during early steps of the method. Adversarially training models using SPGD achieves greater adversarial accuracy compared to PGD when holding the number of attack steps constant. The use of SPGD can, therefore, reduce the overhead of adversarial training when utilizing adversarial generation with a smaller number of steps. However, we also prove that SPGD is equivalent to a variant of the PGD ordinarily used for the $l_{\infty}$ threat model. This PGD variant omits the sign function which is ordinarily applied to the gradient. SPGD can, therefore, be performed without explicitly transforming into the frequency domain. Finally, we visualize the perturbations SPGD generates and find they use both high and low-frequency components, which suggests that removing either high-frequency components or low-frequency components is not an effective defense.
* 9 pages, 9 figures, SPAI'20, ACM ASIACCS 2020
Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation
Oct 22, 2018



Abstract:Deep learning models for semantic segmentation of images require large amounts of data. In the medical imaging domain, acquiring sufficient data is a significant challenge. Labeling medical image data requires expert knowledge. Collaboration between institutions could address this challenge, but sharing medical data to a centralized location faces various legal, privacy, technical, and data-ownership challenges, especially among international institutions. In this study, we introduce the first use of federated learning for multi-institutional collaboration, enabling deep learning modeling without sharing patient data. Our quantitative results demonstrate that the performance of federated semantic segmentation models (Dice=0.852) on multimodal brain scans is similar to that of models trained by sharing data (Dice=0.862). We compare federated learning with two alternative collaborative learning methods and find that they fail to match the performance of federated learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge