Sérgio Pereira
OCELOT 2023: Cell Detection from Cell-Tissue Interaction Challenge
Sep 11, 2025Abstract:Pathologists routinely alternate between different magnifications when examining Whole-Slide Images, allowing them to evaluate both broad tissue morphology and intricate cellular details to form comprehensive diagnoses. However, existing deep learning-based cell detection models struggle to replicate these behaviors and learn the interdependent semantics between structures at different magnifications. A key barrier in the field is the lack of datasets with multi-scale overlapping cell and tissue annotations. The OCELOT 2023 challenge was initiated to gather insights from the community to validate the hypothesis that understanding cell and tissue (cell-tissue) interactions is crucial for achieving human-level performance, and to accelerate the research in this field. The challenge dataset includes overlapping cell detection and tissue segmentation annotations from six organs, comprising 673 pairs sourced from 306 The Cancer Genome Atlas (TCGA) Whole-Slide Images with hematoxylin and eosin staining, divided into training, validation, and test subsets. Participants presented models that significantly enhanced the understanding of cell-tissue relationships. Top entries achieved up to a 7.99 increase in F1-score on the test set compared to the baseline cell-only model that did not incorporate cell-tissue relationships. This is a substantial improvement in performance over traditional cell-only detection methods, demonstrating the need for incorporating multi-scale semantics into the models. This paper provides a comparative analysis of the methods used by participants, highlighting innovative strategies implemented in the OCELOT 2023 challenge.
* This is the accepted manuscript of an article published in Medical Image Analysis (Elsevier). The final version is available at: https://doi.org/10.1016/j.media.2025.103751
SCORPION: Addressing Scanner-Induced Variability in Histopathology
Jul 28, 2025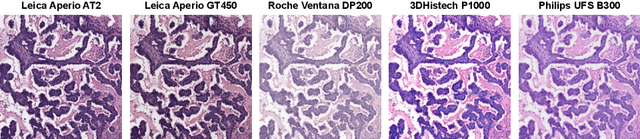
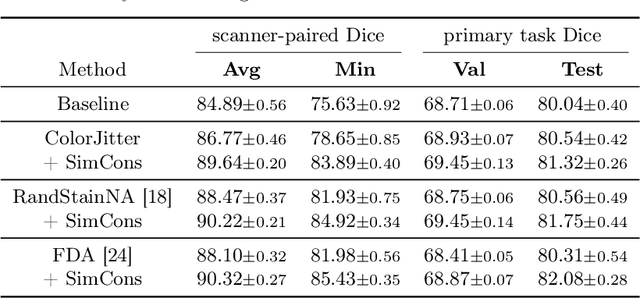


Abstract:Ensuring reliable model performance across diverse domains is a critical challenge in computational pathology. A particular source of variability in Whole-Slide Images is introduced by differences in digital scanners, thus calling for better scanner generalization. This is critical for the real-world adoption of computational pathology, where the scanning devices may differ per institution or hospital, and the model should not be dependent on scanner-induced details, which can ultimately affect the patient's diagnosis and treatment planning. However, past efforts have primarily focused on standard domain generalization settings, evaluating on unseen scanners during training, without directly evaluating consistency across scanners for the same tissue. To overcome this limitation, we introduce SCORPION, a new dataset explicitly designed to evaluate model reliability under scanner variability. SCORPION includes 480 tissue samples, each scanned with 5 scanners, yielding 2,400 spatially aligned patches. This scanner-paired design allows for the isolation of scanner-induced variability, enabling a rigorous evaluation of model consistency while controlling for differences in tissue composition. Furthermore, we propose SimCons, a flexible framework that combines augmentation-based domain generalization techniques with a consistency loss to explicitly address scanner generalization. We empirically show that SimCons improves model consistency on varying scanners without compromising task-specific performance. By releasing the SCORPION dataset and proposing SimCons, we provide the research community with a crucial resource for evaluating and improving model consistency across diverse scanners, setting a new standard for reliability testing.
OCELOT: Overlapped Cell on Tissue Dataset for Histopathology
Mar 24, 2023



Abstract:Cell detection is a fundamental task in computational pathology that can be used for extracting high-level medical information from whole-slide images. For accurate cell detection, pathologists often zoom out to understand the tissue-level structures and zoom in to classify cells based on their morphology and the surrounding context. However, there is a lack of efforts to reflect such behaviors by pathologists in the cell detection models, mainly due to the lack of datasets containing both cell and tissue annotations with overlapping regions. To overcome this limitation, we propose and publicly release OCELOT, a dataset purposely dedicated to the study of cell-tissue relationships for cell detection in histopathology. OCELOT provides overlapping cell and tissue annotations on images acquired from multiple organs. Within this setting, we also propose multi-task learning approaches that benefit from learning both cell and tissue tasks simultaneously. When compared against a model trained only for the cell detection task, our proposed approaches improve cell detection performance on 3 datasets: proposed OCELOT, public TIGER, and internal CARP datasets. On the OCELOT test set in particular, we show up to 6.79 improvement in F1-score. We believe the contributions of this paper, including the release of the OCELOT dataset at https://lunit-io.github.io/research/publications/ocelot are a crucial starting point toward the important research direction of incorporating cell-tissue relationships in computation pathology.
Benchmarking Self-Supervised Learning on Diverse Pathology Datasets
Dec 09, 2022


Abstract:Computational pathology can lead to saving human lives, but models are annotation hungry and pathology images are notoriously expensive to annotate. Self-supervised learning has shown to be an effective method for utilizing unlabeled data, and its application to pathology could greatly benefit its downstream tasks. Yet, there are no principled studies that compare SSL methods and discuss how to adapt them for pathology. To address this need, we execute the largest-scale study of SSL pre-training on pathology image data, to date. Our study is conducted using 4 representative SSL methods on diverse downstream tasks. We establish that large-scale domain-aligned pre-training in pathology consistently out-performs ImageNet pre-training in standard SSL settings such as linear and fine-tuning evaluations, as well as in low-label regimes. Moreover, we propose a set of domain-specific techniques that we experimentally show leads to a performance boost. Lastly, for the first time, we apply SSL to the challenging task of nuclei instance segmentation and show large and consistent performance improvements under diverse settings.
Interactive Multi-Class Tiny-Object Detection
Mar 29, 2022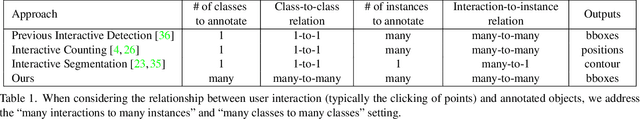

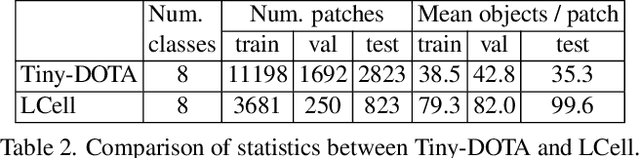
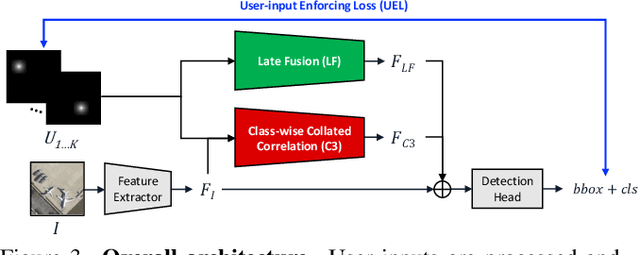
Abstract:Annotating tens or hundreds of tiny objects in a given image is laborious yet crucial for a multitude of Computer Vision tasks. Such imagery typically contains objects from various categories, yet the multi-class interactive annotation setting for the detection task has thus far been unexplored. To address these needs, we propose a novel interactive annotation method for multiple instances of tiny objects from multiple classes, based on a few point-based user inputs. Our approach, C3Det, relates the full image context with annotator inputs in a local and global manner via late-fusion and feature-correlation, respectively. We perform experiments on the Tiny-DOTA and LCell datasets using both two-stage and one-stage object detection architectures to verify the efficacy of our approach. Our approach outperforms existing approaches in interactive annotation, achieving higher mAP with fewer clicks. Furthermore, we validate the annotation efficiency of our approach in a user study where it is shown to be 2.85x faster and yield only 0.36x task load (NASA-TLX, lower is better) compared to manual annotation. The code is available at https://github.com/ChungYi347/Interactive-Multi-Class-Tiny-Object-Detection.
Multi-stage Deep Layer Aggregation for Brain Tumor Segmentation
Jan 02, 2021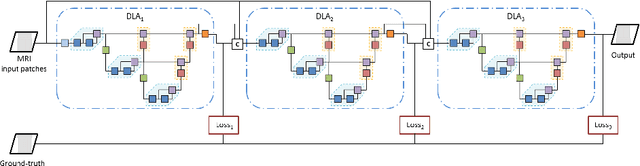
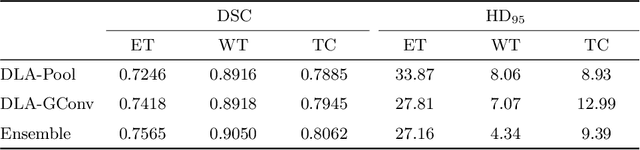
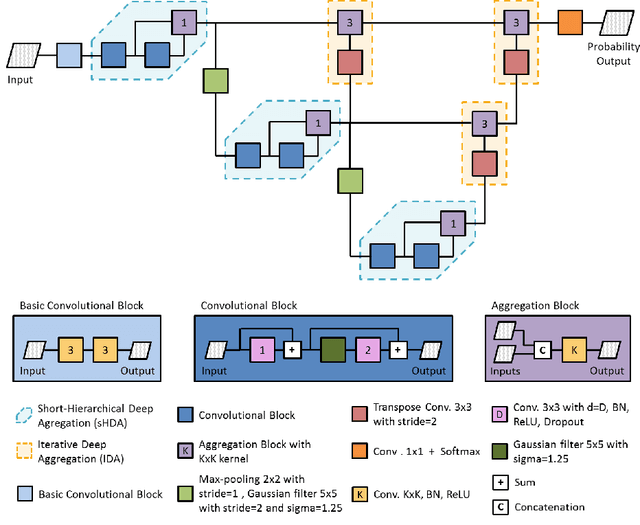
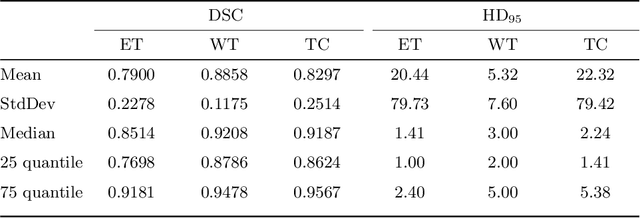
Abstract:Gliomas are among the most aggressive and deadly brain tumors. This paper details the proposed Deep Neural Network architecture for brain tumor segmentation from Magnetic Resonance Images. The architecture consists of a cascade of three Deep Layer Aggregation neural networks, where each stage elaborates the response using the feature maps and the probabilities of the previous stage, and the MRI channels as inputs. The neuroimaging data are part of the publicly available Brain Tumor Segmentation (BraTS) 2020 challenge dataset, where we evaluated our proposal in the BraTS 2020 Validation and Test sets. In the Test set, the experimental results achieved a Dice score of 0.8858, 0.8297 and 0.7900, with an Hausdorff Distance of 5.32 mm, 22.32 mm and 20.44 mm for the whole tumor, core tumor and enhanced tumor, respectively.
Combining unsupervised and supervised learning for predicting the final stroke lesion
Jan 02, 2021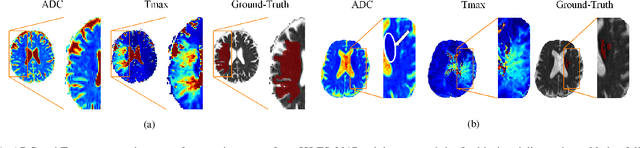
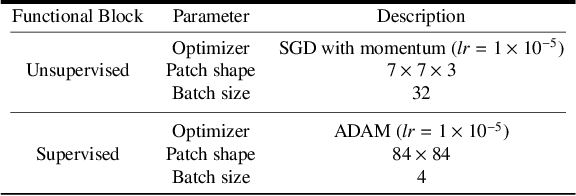
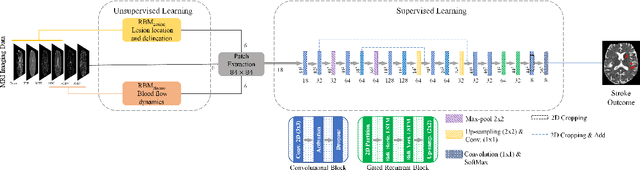
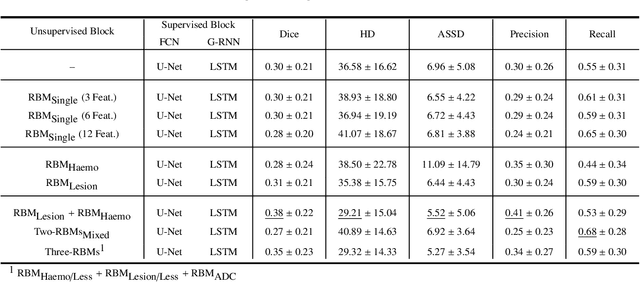
Abstract:Predicting the final ischaemic stroke lesion provides crucial information regarding the volume of salvageable hypoperfused tissue, which helps physicians in the difficult decision-making process of treatment planning and intervention. Treatment selection is influenced by clinical diagnosis, which requires delineating the stroke lesion, as well as characterising cerebral blood flow dynamics using neuroimaging acquisitions. Nonetheless, predicting the final stroke lesion is an intricate task, due to the variability in lesion size, shape, location and the underlying cerebral haemodynamic processes that occur after the ischaemic stroke takes place. Moreover, since elapsed time between stroke and treatment is related to the loss of brain tissue, assessing and predicting the final stroke lesion needs to be performed in a short period of time, which makes the task even more complex. Therefore, there is a need for automatic methods that predict the final stroke lesion and support physicians in the treatment decision process. We propose a fully automatic deep learning method based on unsupervised and supervised learning to predict the final stroke lesion after 90 days. Our aim is to predict the final stroke lesion location and extent, taking into account the underlying cerebral blood flow dynamics that can influence the prediction. To achieve this, we propose a two-branch Restricted Boltzmann Machine, which provides specialized data-driven features from different sets of standard parametric Magnetic Resonance Imaging maps. These data-driven feature maps are then combined with the parametric Magnetic Resonance Imaging maps, and fed to a Convolutional and Recurrent Neural Network architecture. We evaluated our proposal on the publicly available ISLES 2017 testing dataset, reaching a Dice score of 0.38, Hausdorff Distance of 29.21 mm, and Average Symmetric Surface Distance of 5.52 mm.
Retinal vessel segmentation based on Fully Convolutional Neural Networks
Dec 19, 2018



Abstract:The retinal vascular condition is a reliable biomarker of several ophthalmologic and cardiovascular diseases, so automatic vessel segmentation may be crucial to diagnose and monitor them. In this paper, we propose a novel method that combines the multiscale analysis provided by the Stationary Wavelet Transform with a multiscale Fully Convolutional Neural Network to cope with the varying width and direction of the vessel structure in the retina. Our proposal uses rotation operations as the basis of a joint strategy for both data augmentation and prediction, which allows us to explore the information learned during training to refine the segmentation. The method was evaluated on three publicly available databases, achieving an average accuracy of 0.9576, 0.9694, and 0.9653, and average area under the ROC curve of 0.9821, 0.9905, and 0.9855 on the DRIVE, STARE, and CHASE_DB1 databases, respectively. It also appears to be robust to the training set and to the inter-rater variability, which shows its potential for real-world applications.
* Support repository for this work: https://github.com/americofmoliveira/VesselSegmentation_ESWA
Adaptive feature recombination and recalibration for semantic segmentation: application to brain tumor segmentation in MRI
Jun 06, 2018


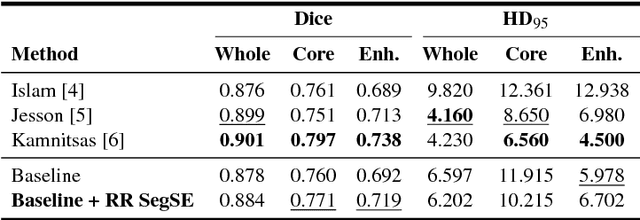
Abstract:Convolutional neural networks (CNNs) have been successfully used for brain tumor segmentation, specifically, fully convolutional networks (FCNs). FCNs can segment a set of voxels at once, having a direct spatial correspondence between units in feature maps (FMs) at a given location and the corresponding classified voxels. In convolutional layers, FMs are merged to create new FMs, so, channel combination is crucial. However, not all FMs have the same relevance for a given class. Recently, in classification problems, Squeeze-and-Excitation (SE) blocks have been proposed to re-calibrate FMs as a whole, and suppress the less informative ones. However, this is not optimal in FCN due to the spatial correspondence between units and voxels. In this article, we propose feature recombination through linear expansion and compression to create more complex features for semantic segmentation. Additionally, we propose a segmentation SE (SegSE) block for feature recalibration that collects contextual information, while maintaining the spatial meaning. Finally, we evaluate the proposed methods in brain tumor segmentation, using publicly available data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge