Wonkyung Jung
OCELOT 2023: Cell Detection from Cell-Tissue Interaction Challenge
Sep 11, 2025Abstract:Pathologists routinely alternate between different magnifications when examining Whole-Slide Images, allowing them to evaluate both broad tissue morphology and intricate cellular details to form comprehensive diagnoses. However, existing deep learning-based cell detection models struggle to replicate these behaviors and learn the interdependent semantics between structures at different magnifications. A key barrier in the field is the lack of datasets with multi-scale overlapping cell and tissue annotations. The OCELOT 2023 challenge was initiated to gather insights from the community to validate the hypothesis that understanding cell and tissue (cell-tissue) interactions is crucial for achieving human-level performance, and to accelerate the research in this field. The challenge dataset includes overlapping cell detection and tissue segmentation annotations from six organs, comprising 673 pairs sourced from 306 The Cancer Genome Atlas (TCGA) Whole-Slide Images with hematoxylin and eosin staining, divided into training, validation, and test subsets. Participants presented models that significantly enhanced the understanding of cell-tissue relationships. Top entries achieved up to a 7.99 increase in F1-score on the test set compared to the baseline cell-only model that did not incorporate cell-tissue relationships. This is a substantial improvement in performance over traditional cell-only detection methods, demonstrating the need for incorporating multi-scale semantics into the models. This paper provides a comparative analysis of the methods used by participants, highlighting innovative strategies implemented in the OCELOT 2023 challenge.
* This is the accepted manuscript of an article published in Medical Image Analysis (Elsevier). The final version is available at: https://doi.org/10.1016/j.media.2025.103751
Generalizing AI-driven Assessment of Immunohistochemistry across Immunostains and Cancer Types: A Universal Immunohistochemistry Analyzer
Jul 30, 2024Abstract:Despite advancements in methodologies, immunohistochemistry (IHC) remains the most utilized ancillary test for histopathologic and companion diagnostics in targeted therapies. However, objective IHC assessment poses challenges. Artificial intelligence (AI) has emerged as a potential solution, yet its development requires extensive training for each cancer and IHC type, limiting versatility. We developed a Universal IHC (UIHC) analyzer, an AI model for interpreting IHC images regardless of tumor or IHC types, using training datasets from various cancers stained for PD-L1 and/or HER2. This multi-cohort trained model outperforms conventional single-cohort models in interpreting unseen IHCs (Kappa score 0.578 vs. up to 0.509) and consistently shows superior performance across different positive staining cutoff values. Qualitative analysis reveals that UIHC effectively clusters patches based on expression levels. The UIHC model also quantitatively assesses c-MET expression with MET mutations, representing a significant advancement in AI application in the era of personalized medicine and accumulating novel biomarkers.
OCELOT: Overlapped Cell on Tissue Dataset for Histopathology
Mar 24, 2023



Abstract:Cell detection is a fundamental task in computational pathology that can be used for extracting high-level medical information from whole-slide images. For accurate cell detection, pathologists often zoom out to understand the tissue-level structures and zoom in to classify cells based on their morphology and the surrounding context. However, there is a lack of efforts to reflect such behaviors by pathologists in the cell detection models, mainly due to the lack of datasets containing both cell and tissue annotations with overlapping regions. To overcome this limitation, we propose and publicly release OCELOT, a dataset purposely dedicated to the study of cell-tissue relationships for cell detection in histopathology. OCELOT provides overlapping cell and tissue annotations on images acquired from multiple organs. Within this setting, we also propose multi-task learning approaches that benefit from learning both cell and tissue tasks simultaneously. When compared against a model trained only for the cell detection task, our proposed approaches improve cell detection performance on 3 datasets: proposed OCELOT, public TIGER, and internal CARP datasets. On the OCELOT test set in particular, we show up to 6.79 improvement in F1-score. We believe the contributions of this paper, including the release of the OCELOT dataset at https://lunit-io.github.io/research/publications/ocelot are a crucial starting point toward the important research direction of incorporating cell-tissue relationships in computation pathology.
AESPA: Accuracy Preserving Low-degree Polynomial Activation for Fast Private Inference
Jan 18, 2022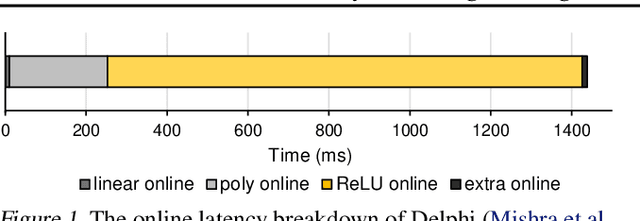
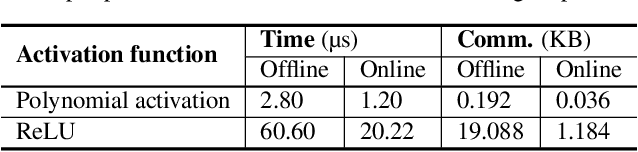
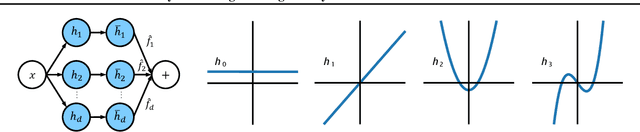

Abstract:Hybrid private inference (PI) protocol, which synergistically utilizes both multi-party computation (MPC) and homomorphic encryption, is one of the most prominent techniques for PI. However, even the state-of-the-art PI protocols are bottlenecked by the non-linear layers, especially the activation functions. Although a standard non-linear activation function can generate higher model accuracy, it must be processed via a costly garbled-circuit MPC primitive. A polynomial activation can be processed via Beaver's multiplication triples MPC primitive but has been incurring severe accuracy drops so far. In this paper, we propose an accuracy preserving low-degree polynomial activation function (AESPA) that exploits the Hermite expansion of the ReLU and basis-wise normalization. We apply AESPA to popular ML models, such as VGGNet, ResNet, and pre-activation ResNet, to show an inference accuracy comparable to those of the standard models with ReLU activation, achieving superior accuracy over prior low-degree polynomial studies. When applied to the all-RELU baseline on the state-of-the-art Delphi PI protocol, AESPA shows up to 42.1x and 28.3x lower online latency and communication cost.
Restructuring Batch Normalization to Accelerate CNN Training
Jul 04, 2018



Abstract:Because CNN models are compute-intensive, where billions of operations can be required just for an inference over a single input image, a variety of CNN accelerators have been proposed and developed. For the early CNN models, the research mostly focused on convolutional and fully-connected layers because the two layers consumed most of the computation cycles. For more recent CNN models, however, non-convolutional layers have become comparably important because of the popular use of newly designed non-convolutional layers and because of the reduction in the number and size of convolutional filters. Non-convolutional layers, including batch normalization (BN), typically have relatively lower computational intensity compared to the convolutional or fully-connected layers, and hence are often constrained by main-memory bandwidth. In this paper, we focus on accelerating the BN layers among the non-convolutional layers, as BN has become a core design block of modern CNNs. A typical modern CNN has a large number of BN layers. BN requires mean and variance calculations over each mini-batch during training. Therefore, the existing memory-access reduction techniques, such as fusing multiple CONV layers, are not effective for accelerating BN due to their inability to optimize mini-batch related calculations. To address this increasingly important problem, we propose to restructure BN layers by first splitting it into two sub-layers and then combining the first sub-layer with its preceding convolutional layer and the second sub-layer with the following activation and convolutional layers. The proposed solution can significantly reduce main-memory accesses while training the latest CNN models, and the experiments on a chip multiprocessor with our modified Caffe implementation show that the proposed BN restructuring can improve the performance of DenseNet with 121 convolutional layers by 28.4%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge