Donggeun Yoo
OCELOT 2023: Cell Detection from Cell-Tissue Interaction Challenge
Sep 11, 2025Abstract:Pathologists routinely alternate between different magnifications when examining Whole-Slide Images, allowing them to evaluate both broad tissue morphology and intricate cellular details to form comprehensive diagnoses. However, existing deep learning-based cell detection models struggle to replicate these behaviors and learn the interdependent semantics between structures at different magnifications. A key barrier in the field is the lack of datasets with multi-scale overlapping cell and tissue annotations. The OCELOT 2023 challenge was initiated to gather insights from the community to validate the hypothesis that understanding cell and tissue (cell-tissue) interactions is crucial for achieving human-level performance, and to accelerate the research in this field. The challenge dataset includes overlapping cell detection and tissue segmentation annotations from six organs, comprising 673 pairs sourced from 306 The Cancer Genome Atlas (TCGA) Whole-Slide Images with hematoxylin and eosin staining, divided into training, validation, and test subsets. Participants presented models that significantly enhanced the understanding of cell-tissue relationships. Top entries achieved up to a 7.99 increase in F1-score on the test set compared to the baseline cell-only model that did not incorporate cell-tissue relationships. This is a substantial improvement in performance over traditional cell-only detection methods, demonstrating the need for incorporating multi-scale semantics into the models. This paper provides a comparative analysis of the methods used by participants, highlighting innovative strategies implemented in the OCELOT 2023 challenge.
* This is the accepted manuscript of an article published in Medical Image Analysis (Elsevier). The final version is available at: https://doi.org/10.1016/j.media.2025.103751
Generalizing AI-driven Assessment of Immunohistochemistry across Immunostains and Cancer Types: A Universal Immunohistochemistry Analyzer
Jul 30, 2024Abstract:Despite advancements in methodologies, immunohistochemistry (IHC) remains the most utilized ancillary test for histopathologic and companion diagnostics in targeted therapies. However, objective IHC assessment poses challenges. Artificial intelligence (AI) has emerged as a potential solution, yet its development requires extensive training for each cancer and IHC type, limiting versatility. We developed a Universal IHC (UIHC) analyzer, an AI model for interpreting IHC images regardless of tumor or IHC types, using training datasets from various cancers stained for PD-L1 and/or HER2. This multi-cohort trained model outperforms conventional single-cohort models in interpreting unseen IHCs (Kappa score 0.578 vs. up to 0.509) and consistently shows superior performance across different positive staining cutoff values. Qualitative analysis reveals that UIHC effectively clusters patches based on expression levels. The UIHC model also quantitatively assesses c-MET expression with MET mutations, representing a significant advancement in AI application in the era of personalized medicine and accumulating novel biomarkers.
Bayesian Optimization Meets Self-Distillation
Apr 25, 2023



Abstract:Bayesian optimization (BO) has contributed greatly to improving model performance by suggesting promising hyperparameter configurations iteratively based on observations from multiple training trials. However, only partial knowledge (i.e., the measured performances of trained models and their hyperparameter configurations) from previous trials is transferred. On the other hand, Self-Distillation (SD) only transfers partial knowledge learned by the task model itself. To fully leverage the various knowledge gained from all training trials, we propose the BOSS framework, which combines BO and SD. BOSS suggests promising hyperparameter configurations through BO and carefully selects pre-trained models from previous trials for SD, which are otherwise abandoned in the conventional BO process. BOSS achieves significantly better performance than both BO and SD in a wide range of tasks including general image classification, learning with noisy labels, semi-supervised learning, and medical image analysis tasks.
OCELOT: Overlapped Cell on Tissue Dataset for Histopathology
Mar 24, 2023



Abstract:Cell detection is a fundamental task in computational pathology that can be used for extracting high-level medical information from whole-slide images. For accurate cell detection, pathologists often zoom out to understand the tissue-level structures and zoom in to classify cells based on their morphology and the surrounding context. However, there is a lack of efforts to reflect such behaviors by pathologists in the cell detection models, mainly due to the lack of datasets containing both cell and tissue annotations with overlapping regions. To overcome this limitation, we propose and publicly release OCELOT, a dataset purposely dedicated to the study of cell-tissue relationships for cell detection in histopathology. OCELOT provides overlapping cell and tissue annotations on images acquired from multiple organs. Within this setting, we also propose multi-task learning approaches that benefit from learning both cell and tissue tasks simultaneously. When compared against a model trained only for the cell detection task, our proposed approaches improve cell detection performance on 3 datasets: proposed OCELOT, public TIGER, and internal CARP datasets. On the OCELOT test set in particular, we show up to 6.79 improvement in F1-score. We believe the contributions of this paper, including the release of the OCELOT dataset at https://lunit-io.github.io/research/publications/ocelot are a crucial starting point toward the important research direction of incorporating cell-tissue relationships in computation pathology.
Benchmarking Self-Supervised Learning on Diverse Pathology Datasets
Dec 09, 2022

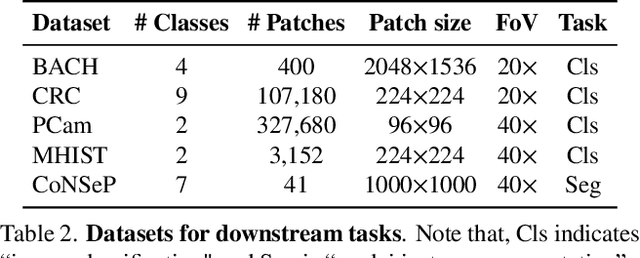

Abstract:Computational pathology can lead to saving human lives, but models are annotation hungry and pathology images are notoriously expensive to annotate. Self-supervised learning has shown to be an effective method for utilizing unlabeled data, and its application to pathology could greatly benefit its downstream tasks. Yet, there are no principled studies that compare SSL methods and discuss how to adapt them for pathology. To address this need, we execute the largest-scale study of SSL pre-training on pathology image data, to date. Our study is conducted using 4 representative SSL methods on diverse downstream tasks. We establish that large-scale domain-aligned pre-training in pathology consistently out-performs ImageNet pre-training in standard SSL settings such as linear and fine-tuning evaluations, as well as in low-label regimes. Moreover, we propose a set of domain-specific techniques that we experimentally show leads to a performance boost. Lastly, for the first time, we apply SSL to the challenging task of nuclei instance segmentation and show large and consistent performance improvements under diverse settings.
Improving Multi-fidelity Optimization with a Recurring Learning Rate for Hyperparameter Tuning
Sep 26, 2022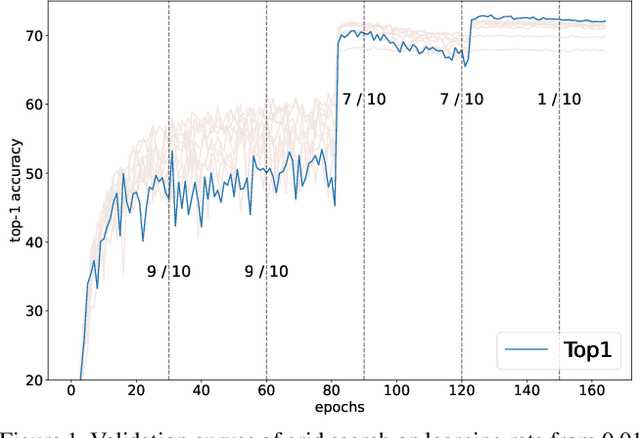
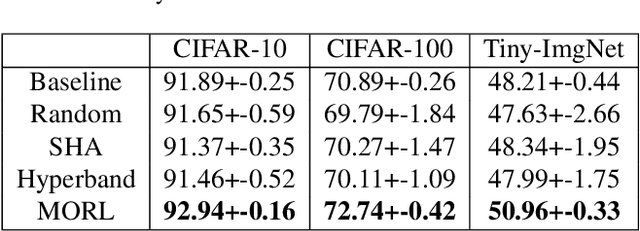
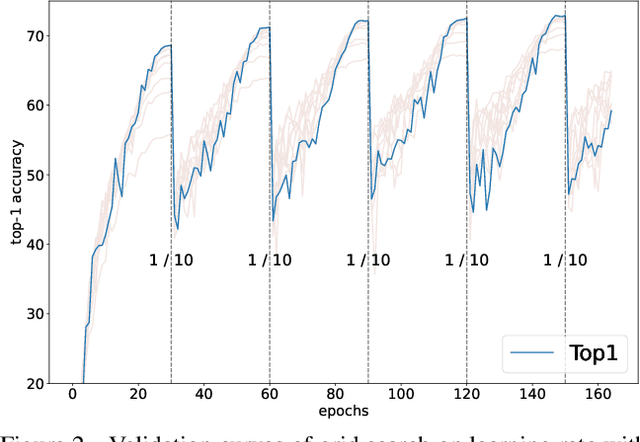
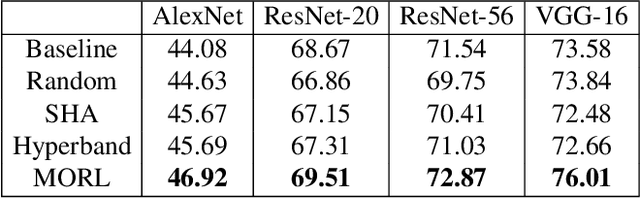
Abstract:Despite the evolution of Convolutional Neural Networks (CNNs), their performance is surprisingly dependent on the choice of hyperparameters. However, it remains challenging to efficiently explore large hyperparameter search space due to the long training times of modern CNNs. Multi-fidelity optimization enables the exploration of more hyperparameter configurations given budget by early termination of unpromising configurations. However, it often results in selecting a sub-optimal configuration as training with the high-performing configuration typically converges slowly in an early phase. In this paper, we propose Multi-fidelity Optimization with a Recurring Learning rate (MORL) which incorporates CNNs' optimization process into multi-fidelity optimization. MORL alleviates the problem of slow-starter and achieves a more precise low-fidelity approximation. Our comprehensive experiments on general image classification, transfer learning, and semi-supervised learning demonstrate the effectiveness of MORL over other multi-fidelity optimization methods such as Successive Halving Algorithm (SHA) and Hyperband. Furthermore, it achieves significant performance improvements over hand-tuned hyperparameter configuration within a practical budget.
Interactive Multi-Class Tiny-Object Detection
Mar 29, 2022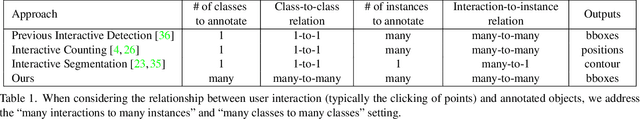

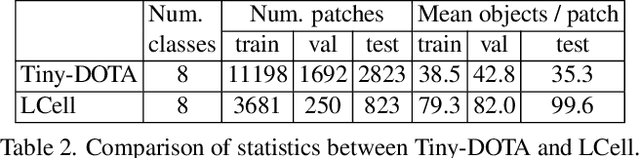
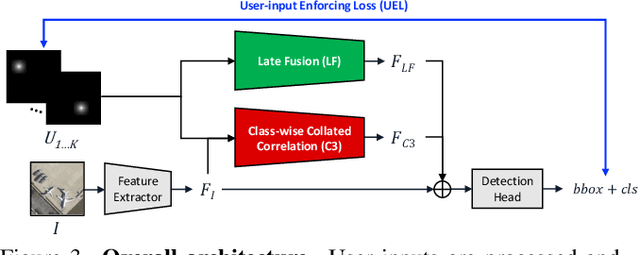
Abstract:Annotating tens or hundreds of tiny objects in a given image is laborious yet crucial for a multitude of Computer Vision tasks. Such imagery typically contains objects from various categories, yet the multi-class interactive annotation setting for the detection task has thus far been unexplored. To address these needs, we propose a novel interactive annotation method for multiple instances of tiny objects from multiple classes, based on a few point-based user inputs. Our approach, C3Det, relates the full image context with annotator inputs in a local and global manner via late-fusion and feature-correlation, respectively. We perform experiments on the Tiny-DOTA and LCell datasets using both two-stage and one-stage object detection architectures to verify the efficacy of our approach. Our approach outperforms existing approaches in interactive annotation, achieving higher mAP with fewer clicks. Furthermore, we validate the annotation efficiency of our approach in a user study where it is shown to be 2.85x faster and yield only 0.36x task load (NASA-TLX, lower is better) compared to manual annotation. The code is available at https://github.com/ChungYi347/Interactive-Multi-Class-Tiny-Object-Detection.
Learning Visual Context by Comparison
Jul 15, 2020



Abstract:Finding diseases from an X-ray image is an important yet highly challenging task. Current methods for solving this task exploit various characteristics of the chest X-ray image, but one of the most important characteristics is still missing: the necessity of comparison between related regions in an image. In this paper, we present Attend-and-Compare Module (ACM) for capturing the difference between an object of interest and its corresponding context. We show that explicit difference modeling can be very helpful in tasks that require direct comparison between locations from afar. This module can be plugged into existing deep learning models. For evaluation, we apply our module to three chest X-ray recognition tasks and COCO object detection & segmentation tasks and observe consistent improvements across tasks. The code is available at https://github.com/mk-minchul/attend-and-compare.
Reducing Domain Gap via Style-Agnostic Networks
Oct 25, 2019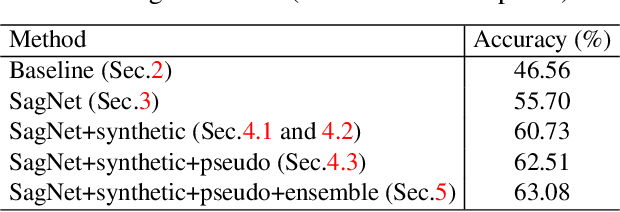
Abstract:Deep learning models often fail to maintain their performance on new test domains. This problem has been regarded as a critical limitation of deep learning for real-world applications. One of the main causes of this vulnerability to domain changes is that the model tends to be biased to image styles (i.e. textures). To tackle this problem, we propose Style-Agnostic Networks (SagNets) to encourage the model to focus more on image contents (i.e. shapes) shared across domains but ignore image styles. SagNets consist of three novel techniques: style adversarial learning, style blending and style consistency learning, each of which prevents the model from making decisions based upon style information. In collaboration with a few additional training techniques and an ensemble of several model variants, the proposed method won the 1st place in the semi-supervised domain adaptation task of the Visual Domain Adaptation 2019 (VisDA-2019) Challenge.
Visuomotor Understanding for Representation Learning of Driving Scenes
Sep 16, 2019



Abstract:Dashboard cameras capture a tremendous amount of driving scene video each day. These videos are purposefully coupled with vehicle sensing data, such as from the speedometer and inertial sensors, providing an additional sensing modality for free. In this work, we leverage the large-scale unlabeled yet naturally paired data for visual representation learning in the driving scenario. A representation is learned in an end-to-end self-supervised framework for predicting dense optical flow from a single frame with paired sensing data. We postulate that success on this task requires the network to learn semantic and geometric knowledge in the ego-centric view. For example, forecasting a future view to be seen from a moving vehicle requires an understanding of scene depth, scale, and movement of objects. We demonstrate that our learned representation can benefit other tasks that require detailed scene understanding and outperforms competing unsupervised representations on semantic segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge