Hyeonsoo Lee
Dual Mixture-of-Experts Framework for Discrete-Time Survival Analysis
Oct 29, 2025Abstract:Survival analysis is a task to model the time until an event of interest occurs, widely used in clinical and biomedical research. A key challenge is to model patient heterogeneity while also adapting risk predictions to both individual characteristics and temporal dynamics. We propose a dual mixture-of-experts (MoE) framework for discrete-time survival analysis. Our approach combines a feature-encoder MoE for subgroup-aware representation learning with a hazard MoE that leverages patient features and time embeddings to capture temporal dynamics. This dual-MoE design flexibly integrates with existing deep learning based survival pipelines. On METABRIC and GBSG breast cancer datasets, our method consistently improves performance, boosting the time-dependent C-index up to 0.04 on the test sets, and yields further gains when incorporated into the Consurv framework.
SelectiveKD: A semi-supervised framework for cancer detection in DBT through Knowledge Distillation and Pseudo-labeling
Sep 25, 2024Abstract:When developing Computer Aided Detection (CAD) systems for Digital Breast Tomosynthesis (DBT), the complexity arising from the volumetric nature of the modality poses significant technical challenges for obtaining large-scale accurate annotations. Without access to large-scale annotations, the resulting model may not generalize to different domains. Given the costly nature of obtaining DBT annotations, how to effectively increase the amount of data used for training DBT CAD systems remains an open challenge. In this paper, we present SelectiveKD, a semi-supervised learning framework for building cancer detection models for DBT, which only requires a limited number of annotated slices to reach high performance. We achieve this by utilizing unlabeled slices available in a DBT stack through a knowledge distillation framework in which the teacher model provides a supervisory signal to the student model for all slices in the DBT volume. Our framework mitigates the potential noise in the supervisory signal from a sub-optimal teacher by implementing a selective dataset expansion strategy using pseudo labels. We evaluate our approach with a large-scale real-world dataset of over 10,000 DBT exams collected from multiple device manufacturers and locations. The resulting SelectiveKD process effectively utilizes unannotated slices from a DBT stack, leading to significantly improved cancer classification performance (AUC) and generalization performance.
Bayesian Optimization Meets Self-Distillation
Apr 25, 2023



Abstract:Bayesian optimization (BO) has contributed greatly to improving model performance by suggesting promising hyperparameter configurations iteratively based on observations from multiple training trials. However, only partial knowledge (i.e., the measured performances of trained models and their hyperparameter configurations) from previous trials is transferred. On the other hand, Self-Distillation (SD) only transfers partial knowledge learned by the task model itself. To fully leverage the various knowledge gained from all training trials, we propose the BOSS framework, which combines BO and SD. BOSS suggests promising hyperparameter configurations through BO and carefully selects pre-trained models from previous trials for SD, which are otherwise abandoned in the conventional BO process. BOSS achieves significantly better performance than both BO and SD in a wide range of tasks including general image classification, learning with noisy labels, semi-supervised learning, and medical image analysis tasks.
Enhancing Breast Cancer Risk Prediction by Incorporating Prior Images
Mar 28, 2023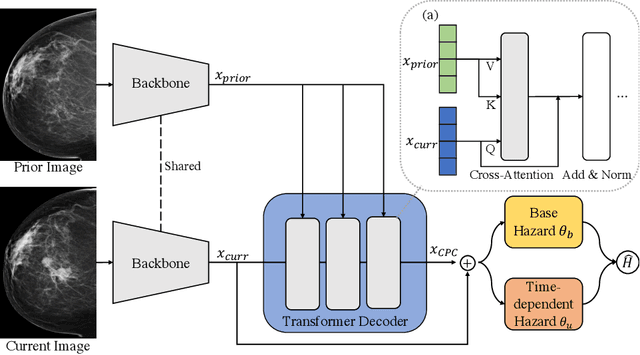
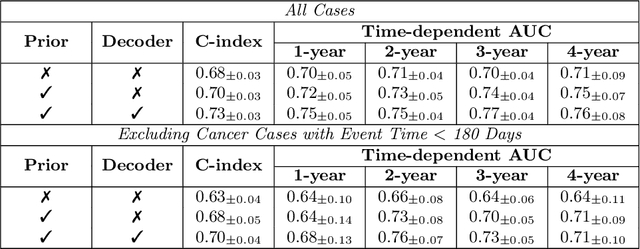
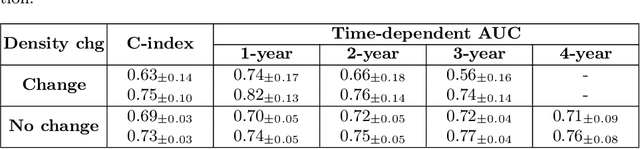
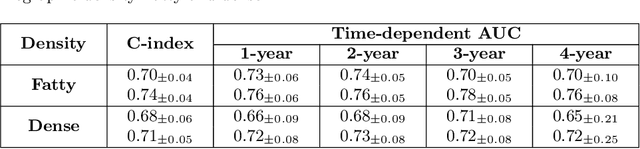
Abstract:Recently, deep learning models have shown the potential to predict breast cancer risk and enable targeted screening strategies, but current models do not consider the change in the breast over time. In this paper, we present a new method, PRIME+, for breast cancer risk prediction that leverages prior mammograms using a transformer decoder, outperforming a state-of-the-art risk prediction method that only uses mammograms from a single time point. We validate our approach on a dataset with 16,113 exams and further demonstrate that it effectively captures patterns of changes from prior mammograms, such as changes in breast density, resulting in improved short-term and long-term breast cancer risk prediction. Experimental results show that our model achieves a statistically significant improvement in performance over the state-of-the-art based model, with a C-index increase from 0.68 to 0.73 (p < 0.05) on held-out test sets.
Scribble2Label: Scribble-Supervised Cell Segmentation via Self-Generating Pseudo-Labels with Consistency
Jun 24, 2020



Abstract:Segmentation is a fundamental process in microscopic cell image analysis. With the advent of recent advances in deep learning, more accurate and high-throughput cell segmentation has become feasible. However, most existing deep learning-based cell segmentation algorithms require fully annotated ground-truth cell labels, which are time-consuming and labor-intensive to generate. In this paper, we introduce Scribble2Label, a novel weakly-supervised cell segmentation framework that exploits only a handful of scribble annotations without full segmentation labels. The core idea is to combine pseudo-labeling and label filtering to generate reliable labels from weak supervision. For this, we leverage the consistency of predictions by iteratively averaging the predictions to improve pseudo labels. We demonstrate the performance of Scribble2Label by comparing it to several state-of-the-art cell segmentation methods with various cell image modalities, including bright-field, fluorescence, and electron microscopy. We also show that our method performs robustly across different levels of scribble details, which confirms that only a few scribble annotations are required in real-use cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge