Thijs Kooi
MV-MLM: Bridging Multi-View Mammography and Language for Breast Cancer Diagnosis and Risk Prediction
Oct 30, 2025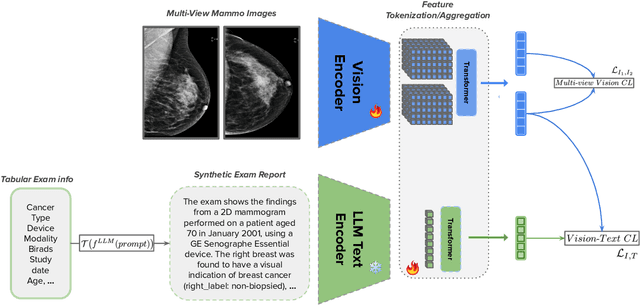
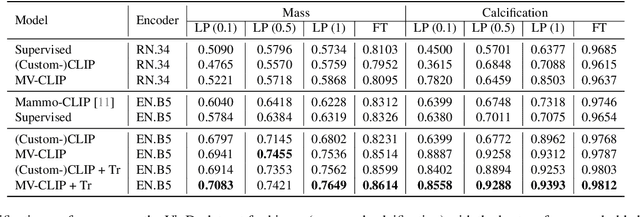


Abstract:Large annotated datasets are essential for training robust Computer-Aided Diagnosis (CAD) models for breast cancer detection or risk prediction. However, acquiring such datasets with fine-detailed annotation is both costly and time-consuming. Vision-Language Models (VLMs), such as CLIP, which are pre-trained on large image-text pairs, offer a promising solution by enhancing robustness and data efficiency in medical imaging tasks. This paper introduces a novel Multi-View Mammography and Language Model for breast cancer classification and risk prediction, trained on a dataset of paired mammogram images and synthetic radiology reports. Our MV-MLM leverages multi-view supervision to learn rich representations from extensive radiology data by employing cross-modal self-supervision across image-text pairs. This includes multiple views and the corresponding pseudo-radiology reports. We propose a novel joint visual-textual learning strategy to enhance generalization and accuracy performance over different data types and tasks to distinguish breast tissues or cancer characteristics(calcification, mass) and utilize these patterns to understand mammography images and predict cancer risk. We evaluated our method on both private and publicly available datasets, demonstrating that the proposed model achieves state-of-the-art performance in three classification tasks: (1) malignancy classification, (2) subtype classification, and (3) image-based cancer risk prediction. Furthermore, the model exhibits strong data efficiency, outperforming existing fully supervised or VLM baselines while trained on synthetic text reports and without the need for actual radiology reports.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
Positive-Sum Fairness: Leveraging Demographic Attributes to Achieve Fair AI Outcomes Without Sacrificing Group Gains
Sep 30, 2024Abstract:Fairness in medical AI is increasingly recognized as a crucial aspect of healthcare delivery. While most of the prior work done on fairness emphasizes the importance of equal performance, we argue that decreases in fairness can be either harmful or non-harmful, depending on the type of change and how sensitive attributes are used. To this end, we introduce the notion of positive-sum fairness, which states that an increase in performance that results in a larger group disparity is acceptable as long as it does not come at the cost of individual subgroup performance. This allows sensitive attributes correlated with the disease to be used to increase performance without compromising on fairness. We illustrate this idea by comparing four CNN models that make different use of the race attribute in the training phase. The results show that removing all demographic encodings from the images helps close the gap in performance between the different subgroups, whereas leveraging the race attribute as a model's input increases the overall performance while widening the disparities between subgroups. These larger gaps are then put in perspective of the collective benefit through our notion of positive-sum fairness to distinguish harmful from non harmful disparities.
SelectiveKD: A semi-supervised framework for cancer detection in DBT through Knowledge Distillation and Pseudo-labeling
Sep 25, 2024Abstract:When developing Computer Aided Detection (CAD) systems for Digital Breast Tomosynthesis (DBT), the complexity arising from the volumetric nature of the modality poses significant technical challenges for obtaining large-scale accurate annotations. Without access to large-scale annotations, the resulting model may not generalize to different domains. Given the costly nature of obtaining DBT annotations, how to effectively increase the amount of data used for training DBT CAD systems remains an open challenge. In this paper, we present SelectiveKD, a semi-supervised learning framework for building cancer detection models for DBT, which only requires a limited number of annotated slices to reach high performance. We achieve this by utilizing unlabeled slices available in a DBT stack through a knowledge distillation framework in which the teacher model provides a supervisory signal to the student model for all slices in the DBT volume. Our framework mitigates the potential noise in the supervisory signal from a sub-optimal teacher by implementing a selective dataset expansion strategy using pseudo labels. We evaluate our approach with a large-scale real-world dataset of over 10,000 DBT exams collected from multiple device manufacturers and locations. The resulting SelectiveKD process effectively utilizes unannotated slices from a DBT stack, leading to significantly improved cancer classification performance (AUC) and generalization performance.
Is user feedback always informative? Retrieval Latent Defending for Semi-Supervised Domain Adaptation without Source Data
Jul 22, 2024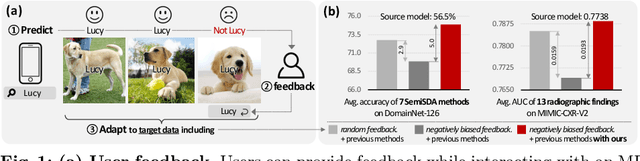
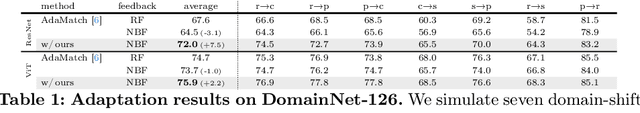
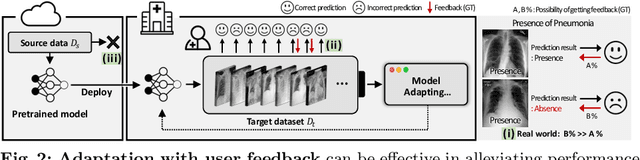
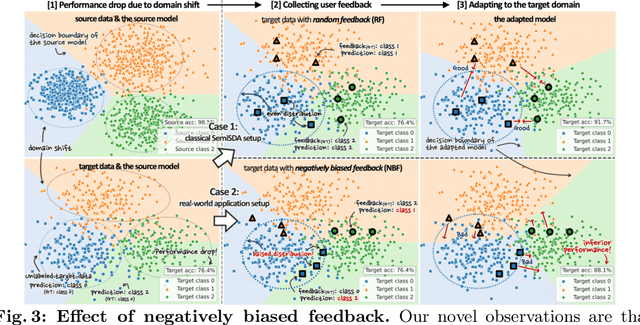
Abstract:This paper aims to adapt the source model to the target environment, leveraging small user feedback (i.e., labeled target data) readily available in real-world applications. We find that existing semi-supervised domain adaptation (SemiSDA) methods often suffer from poorly improved adaptation performance when directly utilizing such feedback data, as shown in Figure 1. We analyze this phenomenon via a novel concept called Negatively Biased Feedback (NBF), which stems from the observation that user feedback is more likely for data points where the model produces incorrect predictions. To leverage this feedback while avoiding the issue, we propose a scalable adapting approach, Retrieval Latent Defending. This approach helps existing SemiSDA methods to adapt the model with a balanced supervised signal by utilizing latent defending samples throughout the adaptation process. We demonstrate the problem caused by NBF and the efficacy of our approach across various benchmarks, including image classification, semantic segmentation, and a real-world medical imaging application. Our extensive experiments reveal that integrating our approach with multiple state-of-the-art SemiSDA methods leads to significant performance improvements.
ELVIS: Empowering Locality of Vision Language Pre-training with Intra-modal Similarity
Apr 11, 2023



Abstract:Deep learning has shown great potential in assisting radiologists in reading chest X-ray (CXR) images, but its need for expensive annotations for improving performance prevents widespread clinical application. Visual language pre-training (VLP) can alleviate the burden and cost of annotation by leveraging routinely generated reports for radiographs, which exist in large quantities as well as in paired form (imagetext pairs). Additionally, extensions to localization-aware VLPs are being proposed to address the needs of accurate localization of abnormalities for CAD in CXR. However, we find that the formulation proposed by locality-aware VLP literatures actually leads to loss in spatial relationships required for downstream localization tasks. Therefore, we propose Empowering Locality of VLP with Intra-modal Similarity, ELVIS, a VLP aware of intra-modal locality, to better preserve the locality within radiographs or reports, which enhances the ability to comprehend location references in text reports. Our locality-aware VLP method significantly outperforms state-of-the art baselines in multiple segmentation tasks and the MS-CXR phrase grounding task. Qualitatively, ELVIS is able to focus well on regions of interest described in the report text compared to prior approaches, allowing for enhanced interpretability.
Enhancing Breast Cancer Risk Prediction by Incorporating Prior Images
Mar 28, 2023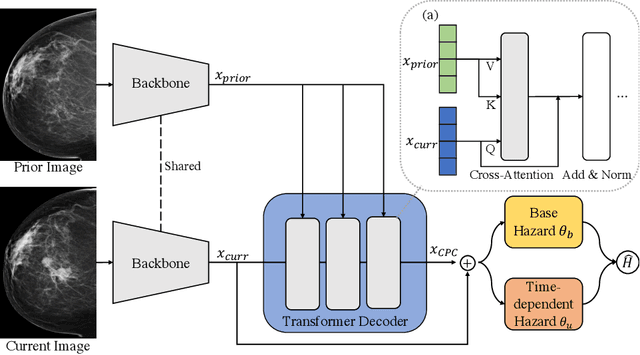
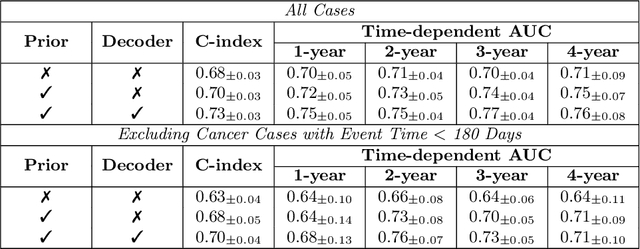
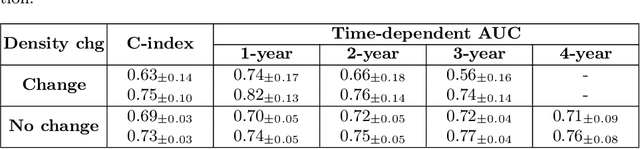
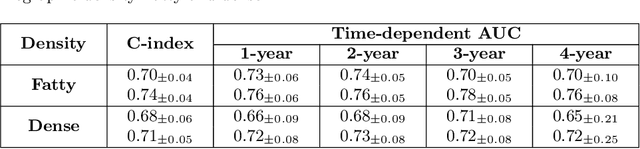
Abstract:Recently, deep learning models have shown the potential to predict breast cancer risk and enable targeted screening strategies, but current models do not consider the change in the breast over time. In this paper, we present a new method, PRIME+, for breast cancer risk prediction that leverages prior mammograms using a transformer decoder, outperforming a state-of-the-art risk prediction method that only uses mammograms from a single time point. We validate our approach on a dataset with 16,113 exams and further demonstrate that it effectively captures patterns of changes from prior mammograms, such as changes in breast density, resulting in improved short-term and long-term breast cancer risk prediction. Experimental results show that our model achieves a statistically significant improvement in performance over the state-of-the-art based model, with a C-index increase from 0.68 to 0.73 (p < 0.05) on held-out test sets.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
OOOE: Only-One-Object-Exists Assumption to Find Very Small Objects in Chest Radiographs
Oct 13, 2022Abstract:The accurate localization of inserted medical tubes and parts of human anatomy is a common problem when analyzing chest radiographs and something deep neural networks could potentially automate. However, many foreign objects like tubes and various anatomical structures are small in comparison to the entire chest X-ray, which leads to severely unbalanced data and makes training deep neural networks difficult. In this paper, we present a simple yet effective `Only-One-Object-Exists' (OOOE) assumption to improve the deep network's ability to localize small landmarks in chest radiographs. The OOOE enables us to recast the localization problem as a classification problem and we can replace commonly used continuous regression techniques with a multi-class discrete objective. We validate our approach using a large scale proprietary dataset of over 100K radiographs as well as publicly available RANZCR-CLiP Kaggle Challenge dataset and show that our method consistently outperforms commonly used regression-based detection models as well as commonly used pixel-wise classification methods. Additionally, we find that the method using the OOOE assumption generalizes to multiple detection problems in chest X-rays and the resulting model shows state-of-the-art performance on detecting various tube tips inserted to the patient as well as patient anatomy.
Did You Get What You Paid For? Rethinking Annotation Cost of Deep Learning Based Computer Aided Detection in Chest Radiographs
Sep 30, 2022Abstract:As deep networks require large amounts of accurately labeled training data, a strategy to collect sufficiently large and accurate annotations is as important as innovations in recognition methods. This is especially true for building Computer Aided Detection (CAD) systems for chest X-rays where domain expertise of radiologists is required to annotate the presence and location of abnormalities on X-ray images. However, there lacks concrete evidence that provides guidance on how much resource to allocate for data annotation such that the resulting CAD system reaches desired performance. Without this knowledge, practitioners often fall back to the strategy of collecting as much detail as possible on as much data as possible which is cost inefficient. In this work, we investigate how the cost of data annotation ultimately impacts the CAD model performance on classification and segmentation of chest abnormalities in frontal-view X-ray images. We define the cost of annotation with respect to the following three dimensions: quantity, quality and granularity of labels. Throughout this study, we isolate the impact of each dimension on the resulting CAD model performance on detecting 10 chest abnormalities in X-rays. On a large scale training data with over 120K X-ray images with gold-standard annotations, we find that cost-efficient annotations provide great value when collected in large amounts and lead to competitive performance when compared to models trained with only gold-standard annotations. We also find that combining large amounts of cost efficient annotations with only small amounts of expensive labels leads to competitive CAD models at a much lower cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge