Michela Antonelli
School of Biomedical Engineering and Imaging Science, King's College London, UK, Hawkes Institute, Department of Computer Science, University College London, UK
Performance uncertainty in medical image analysis: a large-scale investigation of confidence intervals
Jan 23, 2026Abstract:Performance uncertainty quantification is essential for reliable validation and eventual clinical translation of medical imaging artificial intelligence (AI). Confidence intervals (CIs) play a central role in this process by indicating how precise a reported performance estimate is. Yet, due to the limited amount of work examining CI behavior in medical imaging, the community remains largely unaware of how many diverse CI methods exist and how they behave in specific settings. The purpose of this study is to close this gap. To this end, we conducted a large-scale empirical analysis across a total of 24 segmentation and classification tasks, using 19 trained models per task group, a broad spectrum of commonly used performance metrics, multiple aggregation strategies, and several widely adopted CI methods. Reliability (coverage) and precision (width) of each CI method were estimated across all settings to characterize their dependence on study characteristics. Our analysis revealed five principal findings: 1) the sample size required for reliable CIs varies from a few dozens to several thousands of cases depending on study parameters; 2) CI behavior is strongly affected by the choice of performance metric; 3) aggregation strategy substantially influences the reliability of CIs, e.g. they require more observations for macro than for micro; 4) the machine learning problem (segmentation versus classification) modulates these effects; 5) different CI methods are not equally reliable and precise depending on the use case. These results form key components for the development of future guidelines on reporting performance uncertainty in medical imaging AI.
Diffusion-Based Quality Control of Medical Image Segmentations across Organs
Nov 12, 2025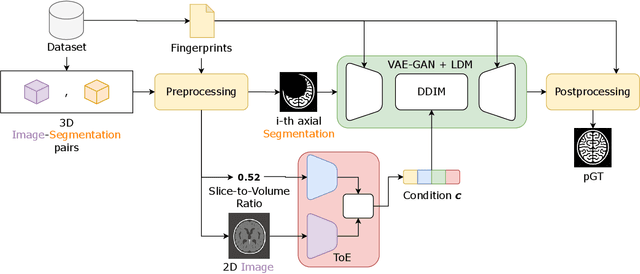
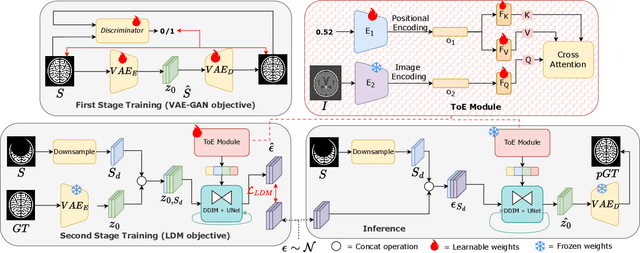
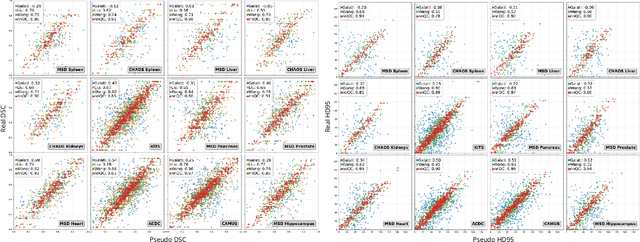
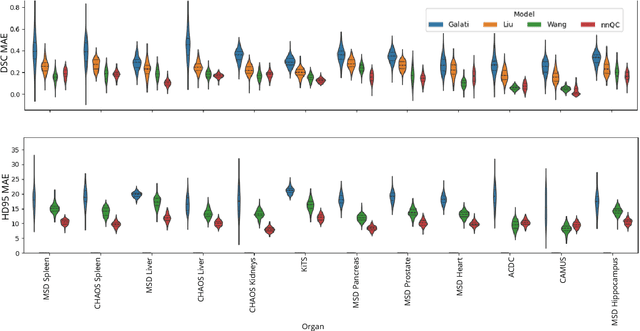
Abstract:Medical image segmentation using deep learning (DL) has enabled the development of automated analysis pipelines for large-scale population studies. However, state-of-the-art DL methods are prone to hallucinations, which can result in anatomically implausible segmentations. With manual correction impractical at scale, automated quality control (QC) techniques have to address the challenge. While promising, existing QC methods are organ-specific, limiting their generalizability and usability beyond their original intended task. To overcome this limitation, we propose no-new Quality Control (nnQC), a robust QC framework based on a diffusion-generative paradigm that self-adapts to any input organ dataset. Central to nnQC is a novel Team of Experts (ToE) architecture, where two specialized experts independently encode 3D spatial awareness, represented by the relative spatial position of an axial slice, and anatomical information derived from visual features from the original image. A weighted conditional module dynamically combines the pair of independent embeddings, or opinions to condition the sampling mechanism within a diffusion process, enabling the generation of a spatially aware pseudo-ground truth for predicting QC scores. Within its framework, nnQC integrates fingerprint adaptation to ensure adaptability across organs, datasets, and imaging modalities. We evaluated nnQC on seven organs using twelve publicly available datasets. Our results demonstrate that nnQC consistently outperforms state-of-the-art methods across all experiments, including cases where segmentation masks are highly degraded or completely missing, confirming its versatility and effectiveness across different organs.
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
Resolution Invariant Autoencoder
Mar 12, 2025Abstract:Deep learning has significantly advanced medical imaging analysis, yet variations in image resolution remain an overlooked challenge. Most methods address this by resampling images, leading to either information loss or computational inefficiencies. While solutions exist for specific tasks, no unified approach has been proposed. We introduce a resolution-invariant autoencoder that adapts spatial resizing at each layer in the network via a learned variable resizing process, replacing fixed spatial down/upsampling at the traditional factor of 2. This ensures a consistent latent space resolution, regardless of input or output resolution. Our model enables various downstream tasks to be performed on an image latent whilst maintaining performance across different resolutions, overcoming the shortfalls of traditional methods. We demonstrate its effectiveness in uncertainty-aware super-resolution, classification, and generative modelling tasks and show how our method outperforms conventional baselines with minimal performance loss across resolutions.
A cautionary tale on the cost-effectiveness of collaborative AI in real-world medical applications
Dec 09, 2024Abstract:Background. Federated learning (FL) has gained wide popularity as a collaborative learning paradigm enabling collaborative AI in sensitive healthcare applications. Nevertheless, the practical implementation of FL presents technical and organizational challenges, as it generally requires complex communication infrastructures. In this context, consensus-based learning (CBL) may represent a promising collaborative learning alternative, thanks to the ability of combining local knowledge into a federated decision system, while potentially reducing deployment overhead. Methods. In this work we propose an extensive benchmark of the accuracy and cost-effectiveness of a panel of FL and CBL methods in a wide range of collaborative medical data analysis scenarios. The benchmark includes 7 different medical datasets, encompassing 3 machine learning tasks, 8 different data modalities, and multi-centric settings involving 3 to 23 clients. Findings. Our results reveal that CBL is a cost-effective alternative to FL. When compared across the panel of medical dataset in the considered benchmark, CBL methods provide equivalent accuracy to the one achieved by FL.Nonetheless, CBL significantly reduces training time and communication cost (resp. 15 fold and 60 fold decrease) (p < 0.05). Interpretation. This study opens a novel perspective on the deployment of collaborative AI in real-world applications, whereas the adoption of cost-effective methods is instrumental to achieve sustainability and democratisation of AI by alleviating the need for extensive computational resources.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024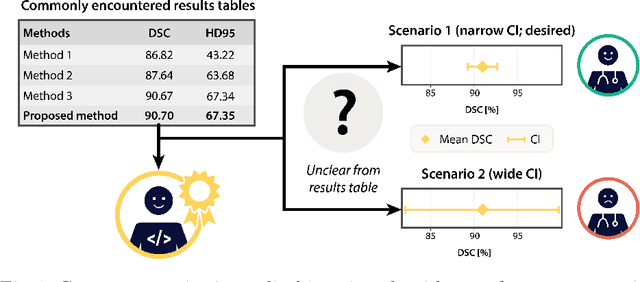
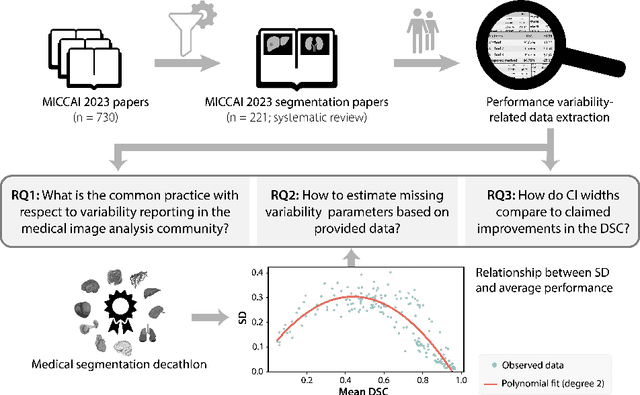
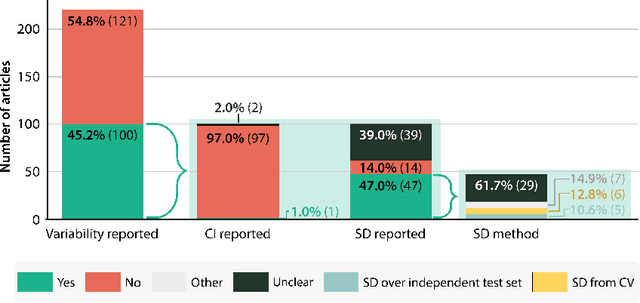

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Benchmarking Collaborative Learning Methods Cost-Effectiveness for Prostate Segmentation
Oct 02, 2023



Abstract:Healthcare data is often split into medium/small-sized collections across multiple hospitals and access to it is encumbered by privacy regulations. This brings difficulties to use them for the development of machine learning and deep learning models, which are known to be data-hungry. One way to overcome this limitation is to use collaborative learning (CL) methods, which allow hospitals to work collaboratively to solve a task, without the need to explicitly share local data. In this paper, we address a prostate segmentation problem from MRI in a collaborative scenario by comparing two different approaches: federated learning (FL) and consensus-based methods (CBM). To the best of our knowledge, this is the first work in which CBM, such as label fusion techniques, are used to solve a problem of collaborative learning. In this setting, CBM combine predictions from locally trained models to obtain a federated strong learner with ideally improved robustness and predictive variance properties. Our experiments show that, in the considered practical scenario, CBMs provide equal or better results than FL, while being highly cost-effective. Our results demonstrate that the consensus paradigm may represent a valid alternative to FL for typical training tasks in medical imaging.
DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images
May 18, 2023Abstract:Automatic segmentation of medical images is a key step for diagnostic and interventional tasks. However, achieving this requires large amounts of annotated volumes, which can be tedious and time-consuming task for expert annotators. In this paper, we introduce DeepEdit, a deep learning-based method for volumetric medical image annotation, that allows automatic and semi-automatic segmentation, and click-based refinement. DeepEdit combines the power of two methods: a non-interactive (i.e. automatic segmentation using nnU-Net, UNET or UNETR) and an interactive segmentation method (i.e. DeepGrow), into a single deep learning model. It allows easy integration of uncertainty-based ranking strategies (i.e. aleatoric and epistemic uncertainty computation) and active learning. We propose and implement a method for training DeepEdit by using standard training combined with user interaction simulation. Once trained, DeepEdit allows clinicians to quickly segment their datasets by using the algorithm in auto segmentation mode or by providing clicks via a user interface (i.e. 3D Slicer, OHIF). We show the value of DeepEdit through evaluation on the PROSTATEx dataset for prostate/prostatic lesions and the Multi-Atlas Labeling Beyond the Cranial Vault (BTCV) dataset for abdominal CT segmentation, using state-of-the-art network architectures as baseline for comparison. DeepEdit could reduce the time and effort annotating 3D medical images compared to DeepGrow alone. Source code is available at https://github.com/Project-MONAI/MONAILabel
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022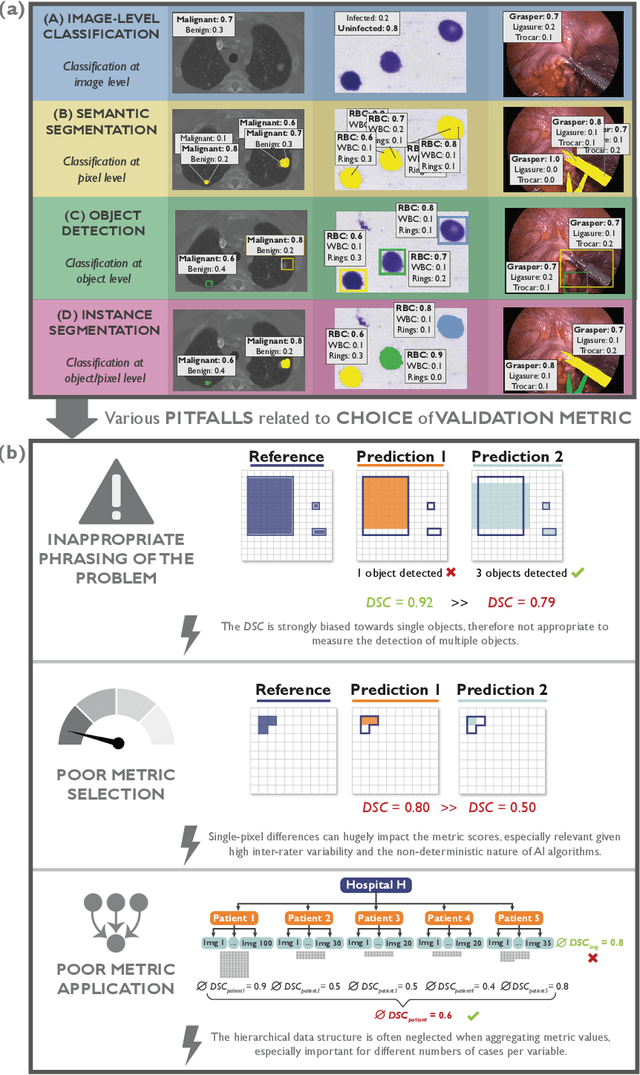
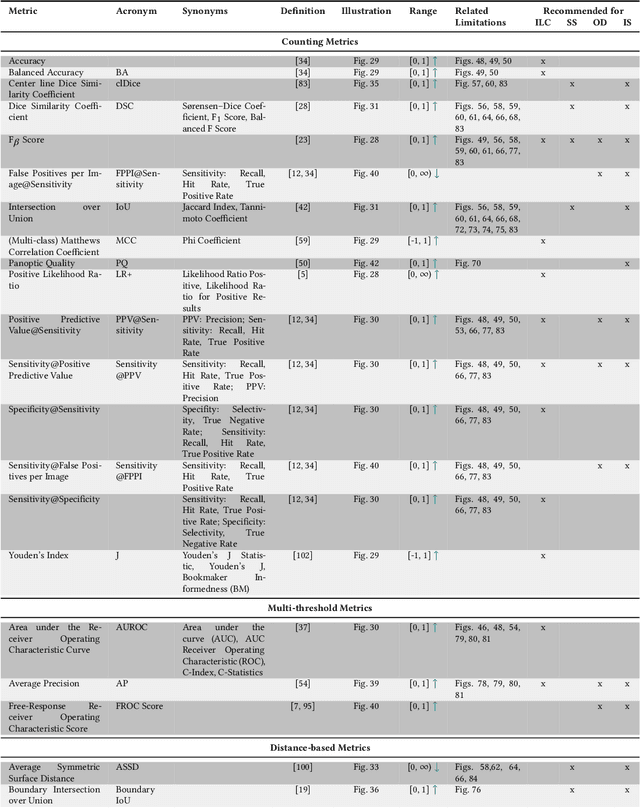
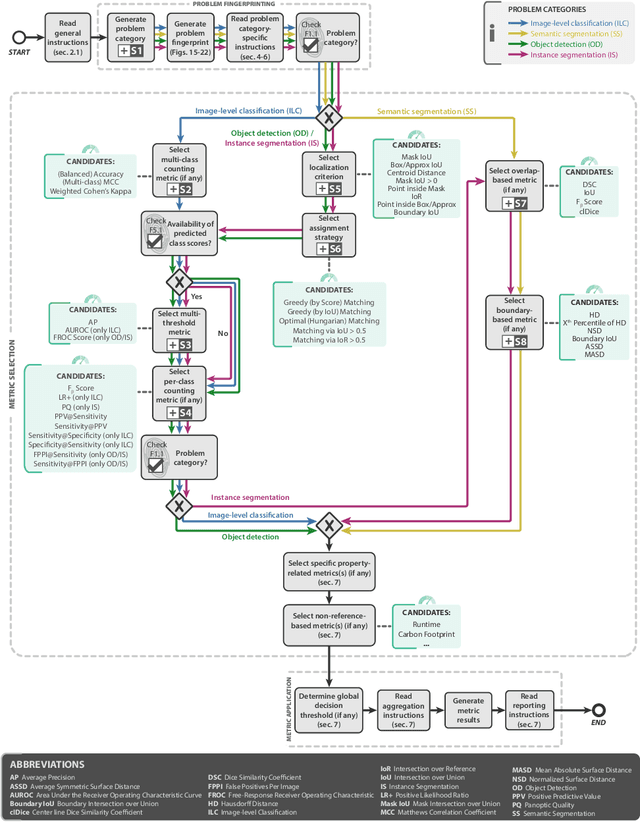
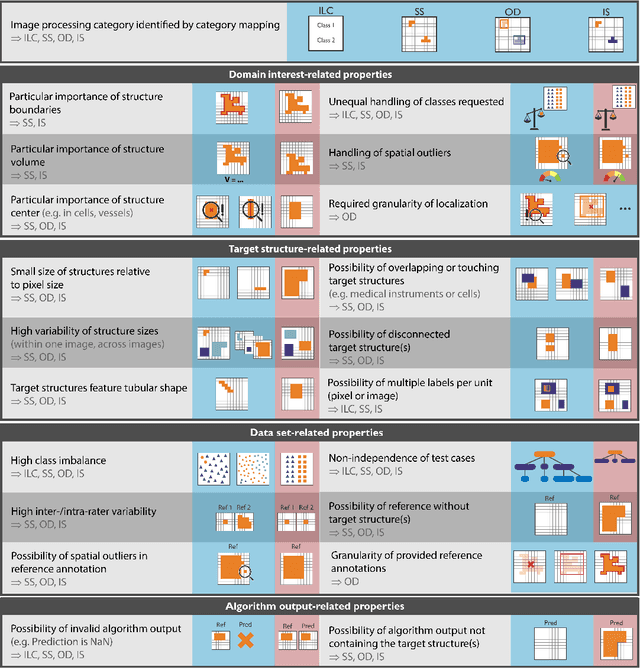
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge