M. Jorge Cardoso
School of Biomedical Engineering and Imaging Science, King's College London, UK
Performance uncertainty in medical image analysis: a large-scale investigation of confidence intervals
Jan 23, 2026Abstract:Performance uncertainty quantification is essential for reliable validation and eventual clinical translation of medical imaging artificial intelligence (AI). Confidence intervals (CIs) play a central role in this process by indicating how precise a reported performance estimate is. Yet, due to the limited amount of work examining CI behavior in medical imaging, the community remains largely unaware of how many diverse CI methods exist and how they behave in specific settings. The purpose of this study is to close this gap. To this end, we conducted a large-scale empirical analysis across a total of 24 segmentation and classification tasks, using 19 trained models per task group, a broad spectrum of commonly used performance metrics, multiple aggregation strategies, and several widely adopted CI methods. Reliability (coverage) and precision (width) of each CI method were estimated across all settings to characterize their dependence on study characteristics. Our analysis revealed five principal findings: 1) the sample size required for reliable CIs varies from a few dozens to several thousands of cases depending on study parameters; 2) CI behavior is strongly affected by the choice of performance metric; 3) aggregation strategy substantially influences the reliability of CIs, e.g. they require more observations for macro than for micro; 4) the machine learning problem (segmentation versus classification) modulates these effects; 5) different CI methods are not equally reliable and precise depending on the use case. These results form key components for the development of future guidelines on reporting performance uncertainty in medical imaging AI.
Diffusion-Based Quality Control of Medical Image Segmentations across Organs
Nov 12, 2025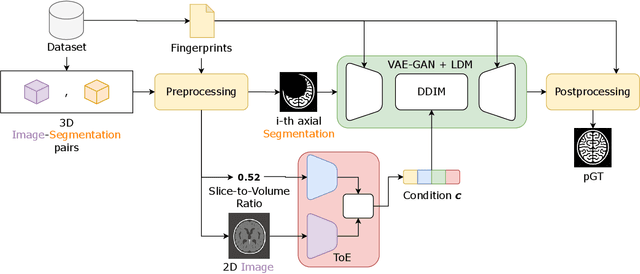
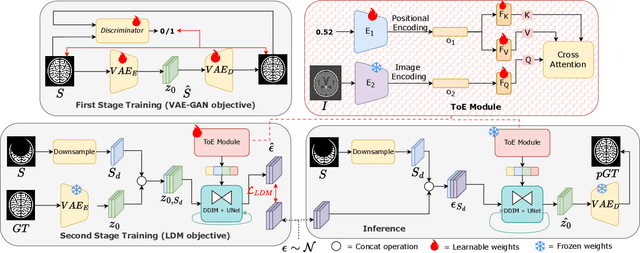
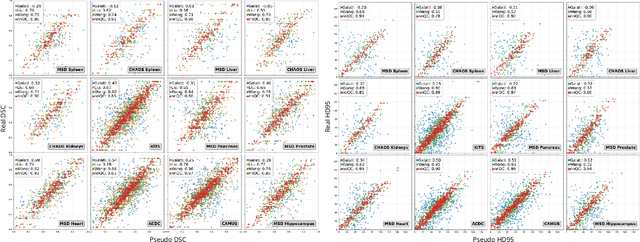
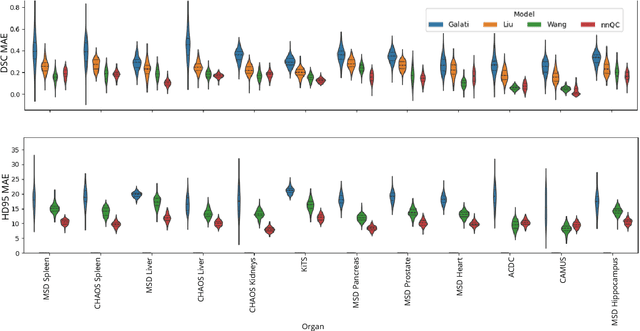
Abstract:Medical image segmentation using deep learning (DL) has enabled the development of automated analysis pipelines for large-scale population studies. However, state-of-the-art DL methods are prone to hallucinations, which can result in anatomically implausible segmentations. With manual correction impractical at scale, automated quality control (QC) techniques have to address the challenge. While promising, existing QC methods are organ-specific, limiting their generalizability and usability beyond their original intended task. To overcome this limitation, we propose no-new Quality Control (nnQC), a robust QC framework based on a diffusion-generative paradigm that self-adapts to any input organ dataset. Central to nnQC is a novel Team of Experts (ToE) architecture, where two specialized experts independently encode 3D spatial awareness, represented by the relative spatial position of an axial slice, and anatomical information derived from visual features from the original image. A weighted conditional module dynamically combines the pair of independent embeddings, or opinions to condition the sampling mechanism within a diffusion process, enabling the generation of a spatially aware pseudo-ground truth for predicting QC scores. Within its framework, nnQC integrates fingerprint adaptation to ensure adaptability across organs, datasets, and imaging modalities. We evaluated nnQC on seven organs using twelve publicly available datasets. Our results demonstrate that nnQC consistently outperforms state-of-the-art methods across all experiments, including cases where segmentation masks are highly degraded or completely missing, confirming its versatility and effectiveness across different organs.
Dynamic causal discovery in Alzheimer's disease through latent pseudotime modelling
Nov 06, 2025



Abstract:The application of causal discovery to diseases like Alzheimer's (AD) is limited by the static graph assumptions of most methods; such models cannot account for an evolving pathophysiology, modulated by a latent disease pseudotime. We propose to apply an existing latent variable model to real-world AD data, inferring a pseudotime that orders patients along a data-driven disease trajectory independent of chronological age, then learning how causal relationships evolve. Pseudotime outperformed age in predicting diagnosis (AUC 0.82 vs 0.59). Incorporating minimal, disease-agnostic background knowledge substantially improved graph accuracy and orientation. Our framework reveals dynamic interactions between novel (NfL, GFAP) and established AD markers, enabling practical causal discovery despite violated assumptions.
A methodology for clinically driven interactive segmentation evaluation
Oct 10, 2025Abstract:Interactive segmentation is a promising strategy for building robust, generalisable algorithms for volumetric medical image segmentation. However, inconsistent and clinically unrealistic evaluation hinders fair comparison and misrepresents real-world performance. We propose a clinically grounded methodology for defining evaluation tasks and metrics, and built a software framework for constructing standardised evaluation pipelines. We evaluate state-of-the-art algorithms across heterogeneous and complex tasks and observe that (i) minimising information loss when processing user interactions is critical for model robustness, (ii) adaptive-zooming mechanisms boost robustness and speed convergence, (iii) performance drops if validation prompting behaviour/budgets differ from training, (iv) 2D methods perform well with slab-like images and coarse targets, but 3D context helps with large or irregularly shaped targets, (v) performance of non-medical-domain models (e.g. SAM2) degrades with poor contrast and complex shapes.
MAIS: Memory-Attention for Interactive Segmentation
May 12, 2025



Abstract:Interactive medical segmentation reduces annotation effort by refining predictions through user feedback. Vision Transformer (ViT)-based models, such as the Segment Anything Model (SAM), achieve state-of-the-art performance using user clicks and prior masks as prompts. However, existing methods treat interactions as independent events, leading to redundant corrections and limited refinement gains. We address this by introducing MAIS, a Memory-Attention mechanism for Interactive Segmentation that stores past user inputs and segmentation states, enabling temporal context integration. Our approach enhances ViT-based segmentation across diverse imaging modalities, achieving more efficient and accurate refinements.
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
Resolution Invariant Autoencoder
Mar 12, 2025Abstract:Deep learning has significantly advanced medical imaging analysis, yet variations in image resolution remain an overlooked challenge. Most methods address this by resampling images, leading to either information loss or computational inefficiencies. While solutions exist for specific tasks, no unified approach has been proposed. We introduce a resolution-invariant autoencoder that adapts spatial resizing at each layer in the network via a learned variable resizing process, replacing fixed spatial down/upsampling at the traditional factor of 2. This ensures a consistent latent space resolution, regardless of input or output resolution. Our model enables various downstream tasks to be performed on an image latent whilst maintaining performance across different resolutions, overcoming the shortfalls of traditional methods. We demonstrate its effectiveness in uncertainty-aware super-resolution, classification, and generative modelling tasks and show how our method outperforms conventional baselines with minimal performance loss across resolutions.
Deep generative computed perfusion-deficit mapping of ischaemic stroke
Feb 03, 2025



Abstract:Focal deficits in ischaemic stroke result from impaired perfusion downstream of a critical vascular occlusion. While parenchymal lesions are traditionally used to predict clinical deficits, the underlying pattern of disrupted perfusion provides information upstream of the lesion, potentially yielding earlier predictive and localizing signals. Such perfusion maps can be derived from routine CT angiography (CTA) widely deployed in clinical practice. Analysing computed perfusion maps from 1,393 CTA-imaged-patients with acute ischaemic stroke, we use deep generative inference to localise neural substrates of NIHSS sub-scores. We show that our approach replicates known lesion-deficit relations without knowledge of the lesion itself and reveals novel neural dependents. The high achieved anatomical fidelity suggests acute CTA-derived computed perfusion maps may be of substantial clinical-and-scientific value in rich phenotyping of acute stroke. Using only hyperacute imaging, deep generative inference could power highly expressive models of functional anatomical relations in ischaemic stroke within the pre-interventional window.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024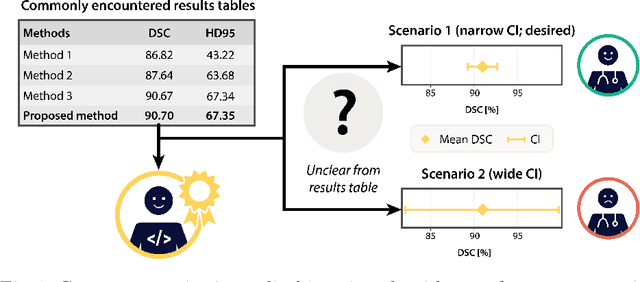
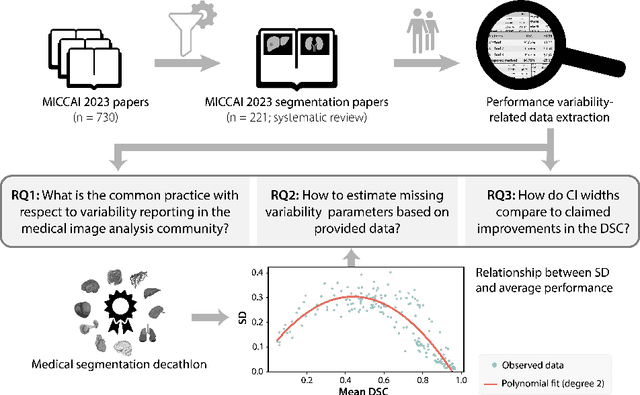
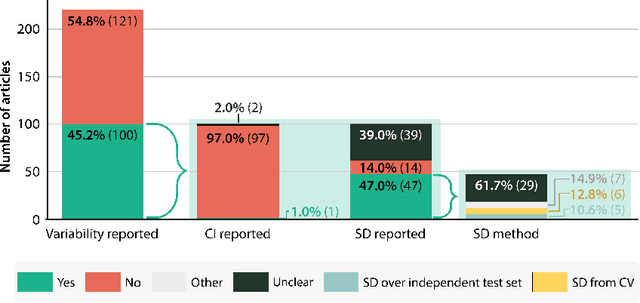

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Framework to generate perfusion map from CT and CTA images in patients with acute ischemic stroke: A longitudinal and cross-sectional study
Apr 05, 2024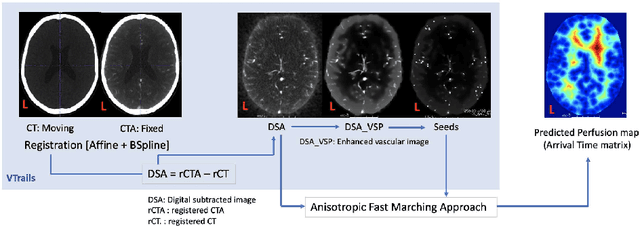
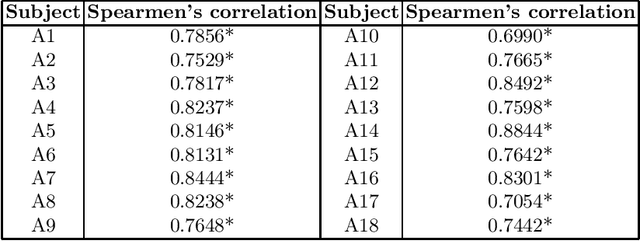
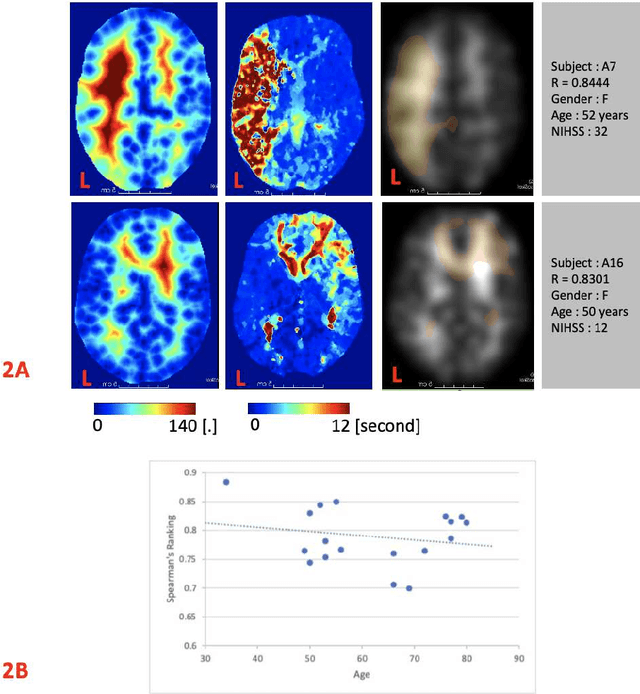
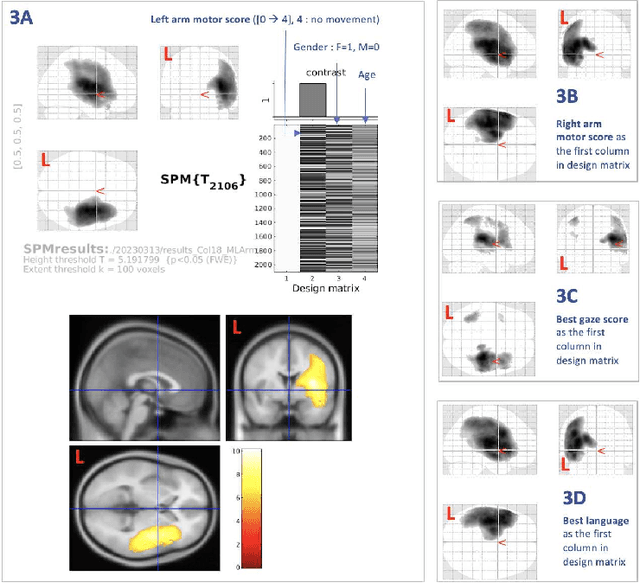
Abstract:Stroke is a leading cause of disability and death. Effective treatment decisions require early and informative vascular imaging. 4D perfusion imaging is ideal but rarely available within the first hour after stroke, whereas plain CT and CTA usually are. Hence, we propose a framework to extract a predicted perfusion map (PPM) derived from CT and CTA images. In all eighteen patients, we found significantly high spatial similarity (with average Spearman's correlation = 0.7893) between our predicted perfusion map (PPM) and the T-max map derived from 4D-CTP. Voxelwise correlations between the PPM and National Institutes of Health Stroke Scale (NIHSS) subscores for L/R hand motor, gaze, and language on a large cohort of 2,110 subjects reliably mapped symptoms to expected infarct locations. Therefore our PPM could serve as an alternative for 4D perfusion imaging, if the latter is unavailable, to investigate blood perfusion in the first hours after hospital admission.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge