Petru-Daniel Tudosiu
King's College London
Exploiting Mixture-of-Experts Redundancy Unlocks Multimodal Generative Abilities
Apr 01, 2025Abstract:In this work, we undertake the challenge of augmenting the existing generative capabilities of pre-trained text-only large language models (LLMs) with multi-modal generation capability while satisfying two core constraints: C1 preserving the preservation of original language generative capabilities with negligible performance degradation, and C2 adhering to a small parameter budget to learn the new modality, ensuring scalability and efficiency. In contrast to current approaches that add dedicated modules, thereby significantly increasing the parameter count, we propose a method that leverages the underutilized capacity inherent in deep models. Specifically, we exploit the parameter redundancy within Mixture-of-Experts (MoEs) as a source of additional capacity for learning a new modality, enabling better parameter efficiency (C1). Moreover, we preserve the original language generation capabilities by applying low-rank adaptation exclusively to the tokens of the new modality (C2). Furthermore, we introduce a novel parameter initialization scheme based on the Gromov-Wasserstein distance to improve convergence and training stability. Through an extensive analysis of the routing mechanism, we uncover the emergence of modality-specific pathways and decreased redundancy within the experts that can efficiently unlock multi-modal generative capabilities. Overall, our method can be seamlessly applied to a wide range of contemporary LLMs, providing a new pathway for transitioning from uni-modal to multi-modal architectures.
Generating Compositional Scenes via Text-to-image RGBA Instance Generation
Nov 16, 2024



Abstract:Text-to-image diffusion generative models can generate high quality images at the cost of tedious prompt engineering. Controllability can be improved by introducing layout conditioning, however existing methods lack layout editing ability and fine-grained control over object attributes. The concept of multi-layer generation holds great potential to address these limitations, however generating image instances concurrently to scene composition limits control over fine-grained object attributes, relative positioning in 3D space and scene manipulation abilities. In this work, we propose a novel multi-stage generation paradigm that is designed for fine-grained control, flexibility and interactivity. To ensure control over instance attributes, we devise a novel training paradigm to adapt a diffusion model to generate isolated scene components as RGBA images with transparency information. To build complex images, we employ these pre-generated instances and introduce a multi-layer composite generation process that smoothly assembles components in realistic scenes. Our experiments show that our RGBA diffusion model is capable of generating diverse and high quality instances with precise control over object attributes. Through multi-layer composition, we demonstrate that our approach allows to build and manipulate images from highly complex prompts with fine-grained control over object appearance and location, granting a higher degree of control than competing methods.
MULAN: A Multi Layer Annotated Dataset for Controllable Text-to-Image Generation
Apr 03, 2024



Abstract:Text-to-image generation has achieved astonishing results, yet precise spatial controllability and prompt fidelity remain highly challenging. This limitation is typically addressed through cumbersome prompt engineering, scene layout conditioning, or image editing techniques which often require hand drawn masks. Nonetheless, pre-existing works struggle to take advantage of the natural instance-level compositionality of scenes due to the typically flat nature of rasterized RGB output images. Towards adressing this challenge, we introduce MuLAn: a novel dataset comprising over 44K MUlti-Layer ANnotations of RGB images as multilayer, instance-wise RGBA decompositions, and over 100K instance images. To build MuLAn, we developed a training free pipeline which decomposes a monocular RGB image into a stack of RGBA layers comprising of background and isolated instances. We achieve this through the use of pretrained general-purpose models, and by developing three modules: image decomposition for instance discovery and extraction, instance completion to reconstruct occluded areas, and image re-assembly. We use our pipeline to create MuLAn-COCO and MuLAn-LAION datasets, which contain a variety of image decompositions in terms of style, composition and complexity. With MuLAn, we provide the first photorealistic resource providing instance decomposition and occlusion information for high quality images, opening up new avenues for text-to-image generative AI research. With this, we aim to encourage the development of novel generation and editing technology, in particular layer-wise solutions. MuLAn data resources are available at https://MuLAn-dataset.github.io/.
Optimisation-Based Multi-Modal Semantic Image Editing
Nov 28, 2023



Abstract:Image editing affords increased control over the aesthetics and content of generated images. Pre-existing works focus predominantly on text-based instructions to achieve desired image modifications, which limit edit precision and accuracy. In this work, we propose an inference-time editing optimisation, designed to extend beyond textual edits to accommodate multiple editing instruction types (e.g. spatial layout-based; pose, scribbles, edge maps). We propose to disentangle the editing task into two competing subtasks: successful local image modifications and global content consistency preservation, where subtasks are guided through two dedicated loss functions. By allowing to adjust the influence of each loss function, we build a flexible editing solution that can be adjusted to user preferences. We evaluate our method using text, pose and scribble edit conditions, and highlight our ability to achieve complex edits, through both qualitative and quantitative experiments.
InverseSR: 3D Brain MRI Super-Resolution Using a Latent Diffusion Model
Aug 23, 2023



Abstract:High-resolution (HR) MRI scans obtained from research-grade medical centers provide precise information about imaged tissues. However, routine clinical MRI scans are typically in low-resolution (LR) and vary greatly in contrast and spatial resolution due to the adjustments of the scanning parameters to the local needs of the medical center. End-to-end deep learning methods for MRI super-resolution (SR) have been proposed, but they require re-training each time there is a shift in the input distribution. To address this issue, we propose a novel approach that leverages a state-of-the-art 3D brain generative model, the latent diffusion model (LDM) trained on UK BioBank, to increase the resolution of clinical MRI scans. The LDM acts as a generative prior, which has the ability to capture the prior distribution of 3D T1-weighted brain MRI. Based on the architecture of the brain LDM, we find that different methods are suitable for different settings of MRI SR, and thus propose two novel strategies: 1) for SR with more sparsity, we invert through both the decoder of the LDM and also through a deterministic Denoising Diffusion Implicit Models (DDIM), an approach we will call InverseSR(LDM); 2) for SR with less sparsity, we invert only through the LDM decoder, an approach we will call InverseSR(Decoder). These two approaches search different latent spaces in the LDM model to find the optimal latent code to map the given LR MRI into HR. The training process of the generative model is independent of the MRI under-sampling process, ensuring the generalization of our method to many MRI SR problems with different input measurements. We validate our method on over 100 brain T1w MRIs from the IXI dataset. Our method can demonstrate that powerful priors given by LDM can be used for MRI reconstruction.
Generative AI for Medical Imaging: extending the MONAI Framework
Jul 27, 2023Abstract:Recent advances in generative AI have brought incredible breakthroughs in several areas, including medical imaging. These generative models have tremendous potential not only to help safely share medical data via synthetic datasets but also to perform an array of diverse applications, such as anomaly detection, image-to-image translation, denoising, and MRI reconstruction. However, due to the complexity of these models, their implementation and reproducibility can be difficult. This complexity can hinder progress, act as a use barrier, and dissuade the comparison of new methods with existing works. In this study, we present MONAI Generative Models, a freely available open-source platform that allows researchers and developers to easily train, evaluate, and deploy generative models and related applications. Our platform reproduces state-of-art studies in a standardised way involving different architectures (such as diffusion models, autoregressive transformers, and GANs), and provides pre-trained models for the community. We have implemented these models in a generalisable fashion, illustrating that their results can be extended to 2D or 3D scenarios, including medical images with different modalities (like CT, MRI, and X-Ray data) and from different anatomical areas. Finally, we adopt a modular and extensible approach, ensuring long-term maintainability and the extension of current applications for future features.
Unsupervised 3D out-of-distribution detection with latent diffusion models
Jul 07, 2023


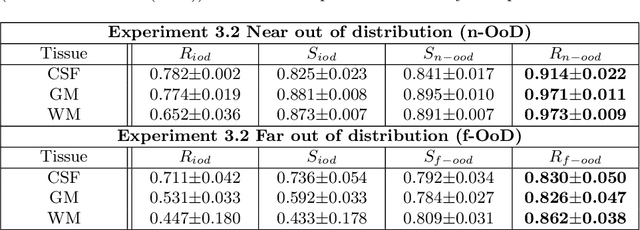
Abstract:Methods for out-of-distribution (OOD) detection that scale to 3D data are crucial components of any real-world clinical deep learning system. Classic denoising diffusion probabilistic models (DDPMs) have been recently proposed as a robust way to perform reconstruction-based OOD detection on 2D datasets, but do not trivially scale to 3D data. In this work, we propose to use Latent Diffusion Models (LDMs), which enable the scaling of DDPMs to high-resolution 3D medical data. We validate the proposed approach on near- and far-OOD datasets and compare it to a recently proposed, 3D-enabled approach using Latent Transformer Models (LTMs). Not only does the proposed LDM-based approach achieve statistically significant better performance, it also shows less sensitivity to the underlying latent representation, more favourable memory scaling, and produces better spatial anomaly maps. Code is available at https://github.com/marksgraham/ddpm-ood
Denoising Diffusion Models for Out-of-Distribution Detection
Nov 30, 2022Abstract:Out-of-distribution detection is crucial to the safe deployment of machine learning systems. Currently, the state-of-the-art in unsupervised out-of-distribution detection is dominated by generative-based approaches that make use of estimates of the likelihood or other measurements from a generative model. Reconstruction-based methods offer an alternative approach, in which a measure of reconstruction error is used to determine if a sample is out-of-distribution. However, reconstruction-based approaches are less favoured, as they require careful tuning of the model's information bottleneck - such as the size of the latent dimension - to produce good results. In this work, we exploit the view of denoising diffusion probabilistic models (DDPM) as denoising autoencoders where the bottleneck is controlled externally, by means of the amount of noise applied. We propose to use DDPMs to reconstruct an input that has been noised to a range of noise levels, and use the resulting multi-dimensional reconstruction error to classify out-of-distribution inputs. Our approach outperforms not only reconstruction-based methods, but also state-of-the-art generative-based approaches.
Can segmentation models be trained with fully synthetically generated data?
Sep 17, 2022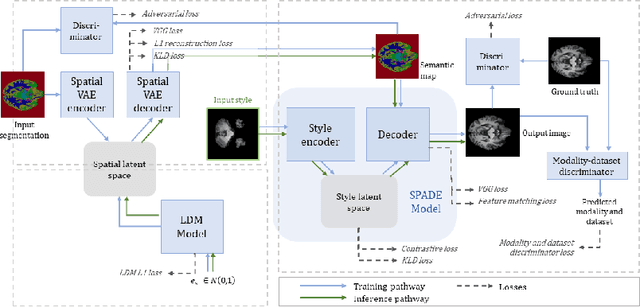

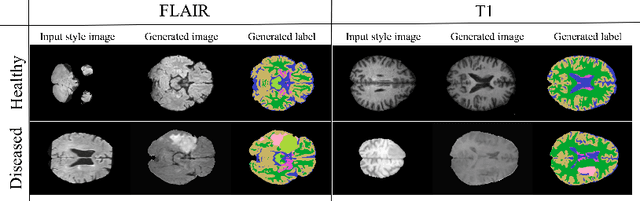
Abstract:In order to achieve good performance and generalisability, medical image segmentation models should be trained on sizeable datasets with sufficient variability. Due to ethics and governance restrictions, and the costs associated with labelling data, scientific development is often stifled, with models trained and tested on limited data. Data augmentation is often used to artificially increase the variability in the data distribution and improve model generalisability. Recent works have explored deep generative models for image synthesis, as such an approach would enable the generation of an effectively infinite amount of varied data, addressing the generalisability and data access problems. However, many proposed solutions limit the user's control over what is generated. In this work, we propose brainSPADE, a model which combines a synthetic diffusion-based label generator with a semantic image generator. Our model can produce fully synthetic brain labels on-demand, with or without pathology of interest, and then generate a corresponding MRI image of an arbitrary guided style. Experiments show that brainSPADE synthetic data can be used to train segmentation models with performance comparable to that of models trained on real data.
Brain Imaging Generation with Latent Diffusion Models
Sep 15, 2022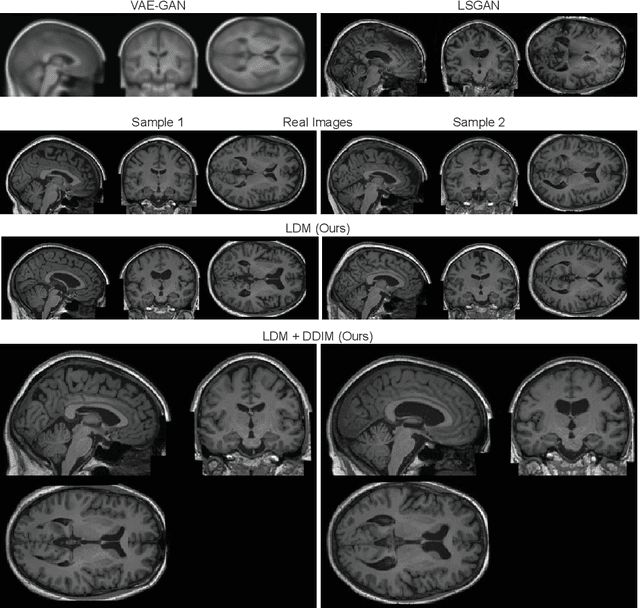
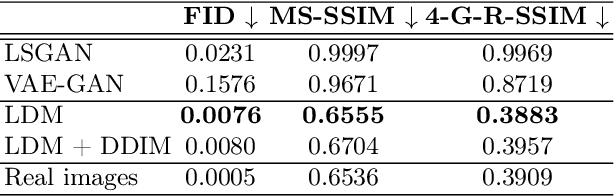
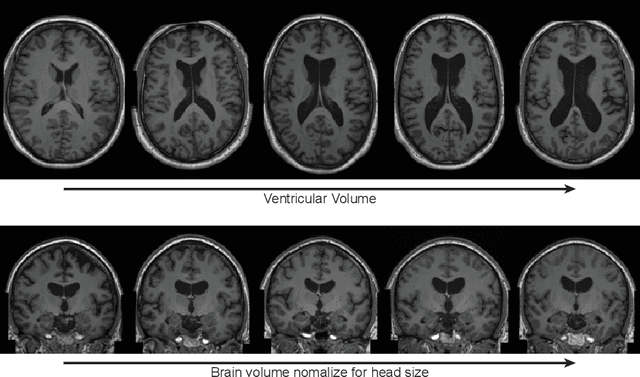
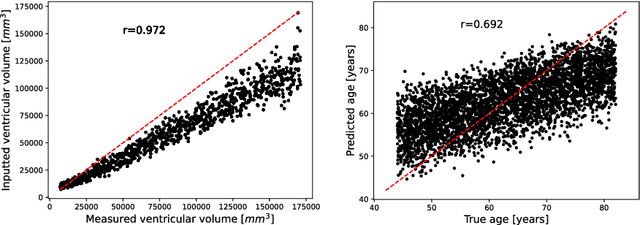
Abstract:Deep neural networks have brought remarkable breakthroughs in medical image analysis. However, due to their data-hungry nature, the modest dataset sizes in medical imaging projects might be hindering their full potential. Generating synthetic data provides a promising alternative, allowing to complement training datasets and conducting medical image research at a larger scale. Diffusion models recently have caught the attention of the computer vision community by producing photorealistic synthetic images. In this study, we explore using Latent Diffusion Models to generate synthetic images from high-resolution 3D brain images. We used T1w MRI images from the UK Biobank dataset (N=31,740) to train our models to learn about the probabilistic distribution of brain images, conditioned on covariables, such as age, sex, and brain structure volumes. We found that our models created realistic data, and we could use the conditioning variables to control the data generation effectively. Besides that, we created a synthetic dataset with 100,000 brain images and made it openly available to the scientific community.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge