Guoxuan Xia
Exploiting Mixture-of-Experts Redundancy Unlocks Multimodal Generative Abilities
Apr 01, 2025Abstract:In this work, we undertake the challenge of augmenting the existing generative capabilities of pre-trained text-only large language models (LLMs) with multi-modal generation capability while satisfying two core constraints: C1 preserving the preservation of original language generative capabilities with negligible performance degradation, and C2 adhering to a small parameter budget to learn the new modality, ensuring scalability and efficiency. In contrast to current approaches that add dedicated modules, thereby significantly increasing the parameter count, we propose a method that leverages the underutilized capacity inherent in deep models. Specifically, we exploit the parameter redundancy within Mixture-of-Experts (MoEs) as a source of additional capacity for learning a new modality, enabling better parameter efficiency (C1). Moreover, we preserve the original language generation capabilities by applying low-rank adaptation exclusively to the tokens of the new modality (C2). Furthermore, we introduce a novel parameter initialization scheme based on the Gromov-Wasserstein distance to improve convergence and training stability. Through an extensive analysis of the routing mechanism, we uncover the emergence of modality-specific pathways and decreased redundancy within the experts that can efficiently unlock multi-modal generative capabilities. Overall, our method can be seamlessly applied to a wide range of contemporary LLMs, providing a new pathway for transitioning from uni-modal to multi-modal architectures.
Generative Uncertainty in Diffusion Models
Feb 28, 2025



Abstract:Diffusion models have recently driven significant breakthroughs in generative modeling. While state-of-the-art models produce high-quality samples on average, individual samples can still be low quality. Detecting such samples without human inspection remains a challenging task. To address this, we propose a Bayesian framework for estimating generative uncertainty of synthetic samples. We outline how to make Bayesian inference practical for large, modern generative models and introduce a new semantic likelihood (evaluated in the latent space of a feature extractor) to address the challenges posed by high-dimensional sample spaces. Through our experiments, we demonstrate that the proposed generative uncertainty effectively identifies poor-quality samples and significantly outperforms existing uncertainty-based methods. Notably, our Bayesian framework can be applied post-hoc to any pretrained diffusion or flow matching model (via the Laplace approximation), and we propose simple yet effective techniques to minimize its computational overhead during sampling.
Towards Understanding and Quantifying Uncertainty for Text-to-Image Generation
Dec 04, 2024Abstract:Uncertainty quantification in text-to-image (T2I) generative models is crucial for understanding model behavior and improving output reliability. In this paper, we are the first to quantify and evaluate the uncertainty of T2I models with respect to the prompt. Alongside adapting existing approaches designed to measure uncertainty in the image space, we also introduce Prompt-based UNCertainty Estimation for T2I models (PUNC), a novel method leveraging Large Vision-Language Models (LVLMs) to better address uncertainties arising from the semantics of the prompt and generated images. PUNC utilizes a LVLM to caption a generated image, and then compares the caption with the original prompt in the more semantically meaningful text space. PUNC also enables the disentanglement of both aleatoric and epistemic uncertainties via precision and recall, which image-space approaches are unable to do. Extensive experiments demonstrate that PUNC outperforms state-of-the-art uncertainty estimation techniques across various settings. Uncertainty quantification in text-to-image generation models can be used on various applications including bias detection, copyright protection, and OOD detection. We also introduce a comprehensive dataset of text prompts and generation pairs to foster further research in uncertainty quantification for generative models. Our findings illustrate that PUNC not only achieves competitive performance but also enables novel applications in evaluating and improving the trustworthiness of text-to-image models.
Absorb & Escape: Overcoming Single Model Limitations in Generating Genomic Sequences
Oct 28, 2024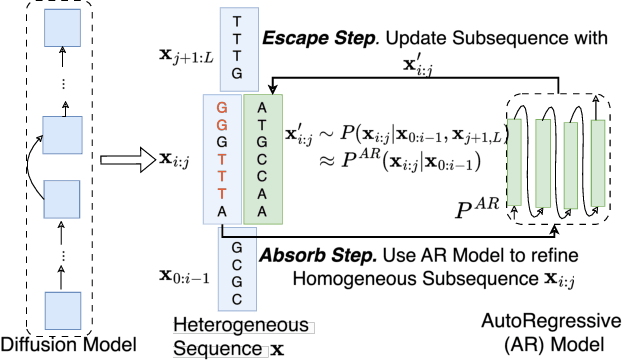

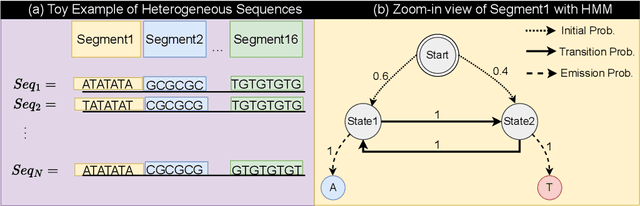

Abstract:Abstract Recent advances in immunology and synthetic biology have accelerated the development of deep generative methods for DNA sequence design. Two dominant approaches in this field are AutoRegressive (AR) models and Diffusion Models (DMs). However, genomic sequences are functionally heterogeneous, consisting of multiple connected regions (e.g., Promoter Regions, Exons, and Introns) where elements within each region come from the same probability distribution, but the overall sequence is non-homogeneous. This heterogeneous nature presents challenges for a single model to accurately generate genomic sequences. In this paper, we analyze the properties of AR models and DMs in heterogeneous genomic sequence generation, pointing out crucial limitations in both methods: (i) AR models capture the underlying distribution of data by factorizing and learning the transition probability but fail to capture the global property of DNA sequences. (ii) DMs learn to recover the global distribution but tend to produce errors at the base pair level. To overcome the limitations of both approaches, we propose a post-training sampling method, termed Absorb & Escape (A&E) to perform compositional generation from AR models and DMs. This approach starts with samples generated by DMs and refines the sample quality using an AR model through the alternation of the Absorb and Escape steps. To assess the quality of generated sequences, we conduct extensive experiments on 15 species for conditional and unconditional DNA generation. The experiment results from motif distribution, diversity checks, and genome integration tests unequivocally show that A&E outperforms state-of-the-art AR models and DMs in genomic sequence generation.
Understanding Why Label Smoothing Degrades Selective Classification and How to Fix It
Mar 19, 2024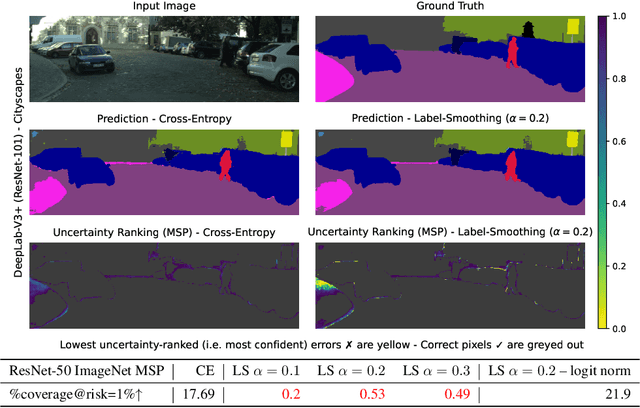
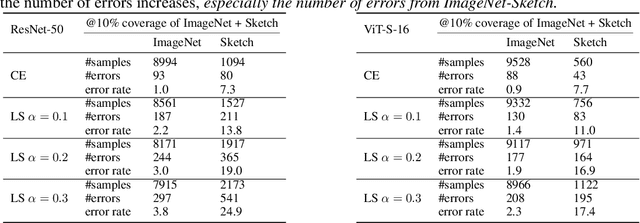
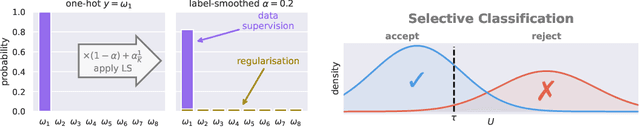

Abstract:Label smoothing (LS) is a popular regularisation method for training deep neural network classifiers due to its effectiveness in improving test accuracy and its simplicity in implementation. "Hard" one-hot labels are "smoothed" by uniformly distributing probability mass to other classes, reducing overfitting. In this work, we reveal that LS negatively affects selective classification (SC) - where the aim is to reject misclassifications using a model's predictive uncertainty. We first demonstrate empirically across a range of tasks and architectures that LS leads to a consistent degradation in SC. We then explain this by analysing logit-level gradients, showing that LS exacerbates overconfidence and underconfidence by regularising the max logit more when the probability of error is low, and less when the probability of error is high. This elucidates previously reported experimental results where strong classifiers underperform in SC. We then demonstrate the empirical effectiveness of logit normalisation for recovering lost SC performance caused by LS. Furthermore, based on our gradient analysis, we explain why such normalisation is effective. We will release our code shortly.
DiscDiff: Latent Diffusion Model for DNA Sequence Generation
Feb 08, 2024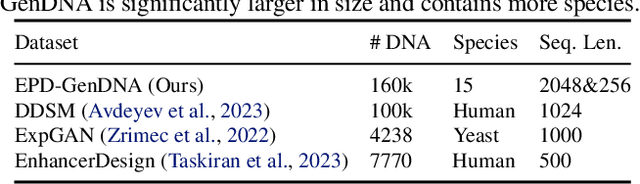
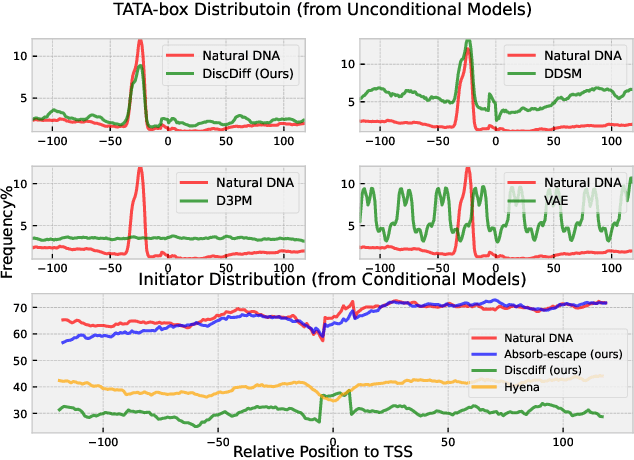

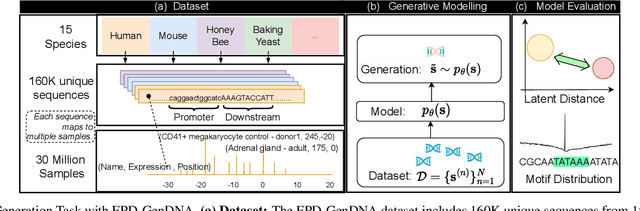
Abstract:This paper introduces a novel framework for DNA sequence generation, comprising two key components: DiscDiff, a Latent Diffusion Model (LDM) tailored for generating discrete DNA sequences, and Absorb-Escape, a post-training algorithm designed to refine these sequences. Absorb-Escape enhances the realism of the generated sequences by correcting `round errors' inherent in the conversion process between latent and input spaces. Our approach not only sets new standards in DNA sequence generation but also demonstrates superior performance over existing diffusion models, in generating both short and long DNA sequences. Additionally, we introduce EPD-GenDNA, the first comprehensive, multi-species dataset for DNA generation, encompassing 160,000 unique sequences from 15 species. We hope this study will advance the generative modelling of DNA, with potential implications for gene therapy and protein production.
Score Normalization for a Faster Diffusion Exponential Integrator Sampler
Nov 10, 2023Abstract:Recently, Zhang et al. have proposed the Diffusion Exponential Integrator Sampler (DEIS) for fast generation of samples from Diffusion Models. It leverages the semi-linear nature of the probability flow ordinary differential equation (ODE) in order to greatly reduce integration error and improve generation quality at low numbers of function evaluations (NFEs). Key to this approach is the score function reparameterisation, which reduces the integration error incurred from using a fixed score function estimate over each integration step. The original authors use the default parameterisation used by models trained for noise prediction -- multiply the score by the standard deviation of the conditional forward noising distribution. We find that although the mean absolute value of this score parameterisation is close to constant for a large portion of the reverse sampling process, it changes rapidly at the end of sampling. As a simple fix, we propose to instead reparameterise the score (at inference) by dividing it by the average absolute value of previous score estimates at that time step collected from offline high NFE generations. We find that our score normalisation (DEIS-SN) consistently improves FID compared to vanilla DEIS, showing an improvement at 10 NFEs from 6.44 to 5.57 on CIFAR-10 and from 5.9 to 4.95 on LSUN-Church 64x64. Our code is available at https://github.com/mtkresearch/Diffusion-DEIS-SN
Latent Diffusion Model for DNA Sequence Generation
Oct 09, 2023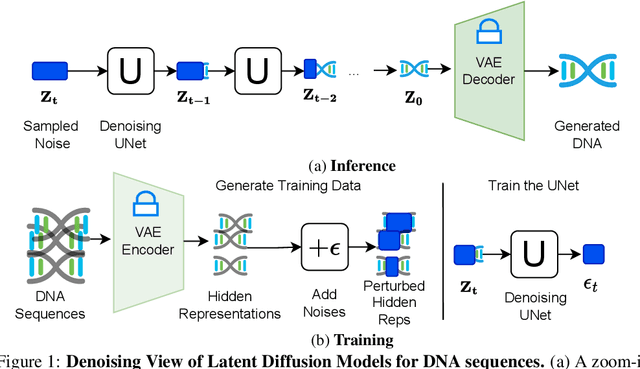
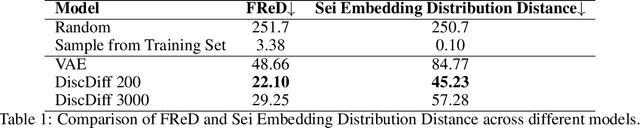
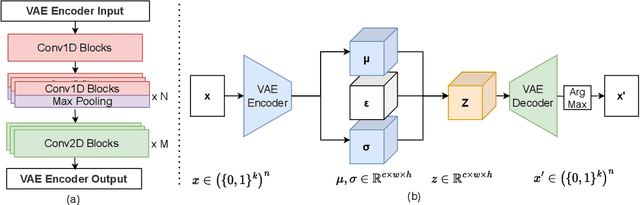
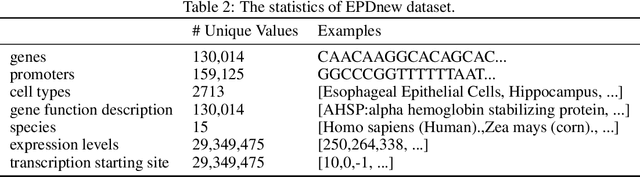
Abstract:The harnessing of machine learning, especially deep generative models, has opened up promising avenues in the field of synthetic DNA sequence generation. Whilst Generative Adversarial Networks (GANs) have gained traction for this application, they often face issues such as limited sample diversity and mode collapse. On the other hand, Diffusion Models are a promising new class of generative models that are not burdened with these problems, enabling them to reach the state-of-the-art in domains such as image generation. In light of this, we propose a novel latent diffusion model, DiscDiff, tailored for discrete DNA sequence generation. By simply embedding discrete DNA sequences into a continuous latent space using an autoencoder, we are able to leverage the powerful generative abilities of continuous diffusion models for the generation of discrete data. Additionally, we introduce Fr\'echet Reconstruction Distance (FReD) as a new metric to measure the sample quality of DNA sequence generations. Our DiscDiff model demonstrates an ability to generate synthetic DNA sequences that align closely with real DNA in terms of Motif Distribution, Latent Embedding Distribution (FReD), and Chromatin Profiles. Additionally, we contribute a comprehensive cross-species dataset of 150K unique promoter-gene sequences from 15 species, enriching resources for future generative modelling in genomics. We will make our code public upon publication.
Logit-Based Ensemble Distribution Distillation for Robust Autoregressive Sequence Uncertainties
May 17, 2023Abstract:Efficiently and reliably estimating uncertainty is an important objective in deep learning. It is especially pertinent to autoregressive sequence tasks, where training and inference costs are typically very high. However, existing research has predominantly focused on tasks with static data such as image classification. In this work, we investigate Ensemble Distribution Distillation (EDD) applied to large-scale natural language sequence-to-sequence data. EDD aims to compress the superior uncertainty performance of an expensive (teacher) ensemble into a cheaper (student) single model. Importantly, the ability to separate knowledge (epistemic) and data (aleatoric) uncertainty is retained. Existing probability-space approaches to EDD, however, are difficult to scale to large vocabularies. We show, for modern transformer architectures on large-scale translation tasks, that modelling the ensemble logits, instead of softmax probabilities, leads to significantly better students. Moreover, the students surprisingly even outperform Deep Ensembles by up to ~10% AUROC on out-of-distribution detection, whilst matching them at in-distribution translation.
Window-Based Early-Exit Cascades for Uncertainty Estimation: When Deep Ensembles are More Efficient than Single Models
Mar 14, 2023



Abstract:Deep Ensembles are a simple, reliable, and effective method of improving both the predictive performance and uncertainty estimates of deep learning approaches. However, they are widely criticised as being computationally expensive, due to the need to deploy multiple independent models. Recent work has challenged this view, showing that for predictive accuracy, ensembles can be more computationally efficient (at inference) than scaling single models within an architecture family. This is achieved by cascading ensemble members via an early-exit approach. In this work, we investigate extending these efficiency gains to tasks related to uncertainty estimation. As many such tasks, e.g. selective classification, are binary classification, our key novel insight is to only pass samples within a window close to the binary decision boundary to later cascade stages. Experiments on ImageNet-scale data across a number of network architectures and uncertainty tasks show that the proposed window-based early-exit approach is able to achieve a superior uncertainty-computation trade-off compared to scaling single models. For example, a cascaded EfficientNet-B2 ensemble is able to achieve similar coverage at 5% risk as a single EfficientNet-B4 with <30% the number of MACs. We also find that cascades/ensembles give more reliable improvements on OOD data vs scaling models up. Code for this work is available at: https://github.com/Guoxoug/window-early-exit.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge