Tom Vercauteren
School of Biomedical Engineering and Imaging Sciences, Kings College London
Learning From a Steady Hand: A Weakly Supervised Agent for Robot Assistance under Microscopy
Jan 28, 2026Abstract:This paper rethinks steady-hand robotic manipulation by using a weakly supervised framework that fuses calibration-aware perception with admittance control. Unlike conventional automation that relies on labor-intensive 2D labeling, our framework leverages reusable warm-up trajectories to extract implicit spatial information, thereby achieving calibration-aware, depth-resolved perception without the need for external fiducials or manual depth annotation. By explicitly characterizing residuals from observation and calibration models, the system establishes a task-space error budget from recorded warm-ups. The uncertainty budget yields a lateral closed-loop accuracy of approx. 49 micrometers at 95% confidence (worst-case testing subset) and a depth accuracy of <= 291 micrometers at 95% confidence bound during large in-plane moves. In a within-subject user study (N=8), the learned agent reduces overall NASA-TLX workload by 77.1% relative to the simple steady-hand assistance baseline. These results demonstrate that the weakly supervised agent improves the reliability of microscope-guided biomedical micromanipulation without introducing complex setup requirements, offering a practical framework for microscope-guided intervention.
Where It Moves, It Matters: Referring Surgical Instrument Segmentation via Motion
Jan 18, 2026Abstract:Enabling intuitive, language-driven interaction with surgical scenes is a critical step toward intelligent operating rooms and autonomous surgical robotic assistance. However, the task of referring segmentation, localizing surgical instruments based on natural language descriptions, remains underexplored in surgical videos, with existing approaches struggling to generalize due to reliance on static visual cues and predefined instrument names. In this work, we introduce SurgRef, a novel motion-guided framework that grounds free-form language expressions in instrument motion, capturing how tools move and interact across time, rather than what they look like. This allows models to understand and segment instruments even under occlusion, ambiguity, or unfamiliar terminology. To train and evaluate SurgRef, we present Ref-IMotion, a diverse, multi-institutional video dataset with dense spatiotemporal masks and rich motion-centric expressions. SurgRef achieves state-of-the-art accuracy and generalization across surgical procedures, setting a new benchmark for robust, language-driven surgical video segmentation.
Beyond one-hot encoding? Journey into compact encoding for large multi-class segmentation
Oct 01, 2025Abstract:This work presents novel methods to reduce computational and memory requirements for medical image segmentation with a large number of classes. We curiously observe challenges in maintaining state-of-the-art segmentation performance with all of the explored options. Standard learning-based methods typically employ one-hot encoding of class labels. The computational complexity and memory requirements thus increase linearly with the number of classes. We propose a family of binary encoding approaches instead of one-hot encoding to reduce the computational complexity and memory requirements to logarithmic in the number of classes. In addition to vanilla binary encoding, we investigate the effects of error-correcting output codes (ECOCs), class weighting, hard/soft decoding, class-to-codeword assignment, and label embedding trees. We apply the methods to the use case of whole brain parcellation with 108 classes based on 3D MRI images. While binary encodings have proven efficient in so-called extreme classification problems in computer vision, we faced challenges in reaching state-of-the-art segmentation quality with binary encodings. Compared to one-hot encoding (Dice Similarity Coefficient (DSC) = 82.4 (2.8)), we report reduced segmentation performance with the binary segmentation approaches, achieving DSCs in the range from 39.3 to 73.8. Informative negative results all too often go unpublished. We hope that this work inspires future research of compact encoding strategies for large multi-class segmentation tasks.
Comparative validation of surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation in endoscopy: Results of the PhaKIR 2024 challenge
Jul 22, 2025Abstract:Reliable recognition and localization of surgical instruments in endoscopic video recordings are foundational for a wide range of applications in computer- and robot-assisted minimally invasive surgery (RAMIS), including surgical training, skill assessment, and autonomous assistance. However, robust performance under real-world conditions remains a significant challenge. Incorporating surgical context - such as the current procedural phase - has emerged as a promising strategy to improve robustness and interpretability. To address these challenges, we organized the Surgical Procedure Phase, Keypoint, and Instrument Recognition (PhaKIR) sub-challenge as part of the Endoscopic Vision (EndoVis) challenge at MICCAI 2024. We introduced a novel, multi-center dataset comprising thirteen full-length laparoscopic cholecystectomy videos collected from three distinct medical institutions, with unified annotations for three interrelated tasks: surgical phase recognition, instrument keypoint estimation, and instrument instance segmentation. Unlike existing datasets, ours enables joint investigation of instrument localization and procedural context within the same data while supporting the integration of temporal information across entire procedures. We report results and findings in accordance with the BIAS guidelines for biomedical image analysis challenges. The PhaKIR sub-challenge advances the field by providing a unique benchmark for developing temporally aware, context-driven methods in RAMIS and offers a high-quality resource to support future research in surgical scene understanding.
Collaborative Human-Robot Surgery for Mandibular Angle Split Osteotomy: Optical Tracking based Approach
Jul 10, 2025Abstract:Mandibular Angle Split Osteotomy (MASO) is a significant procedure in oral and maxillofacial surgery. Despite advances in technique and instrumentation, its success still relies heavily on the surgeon's experience. In this work, a human-robot collaborative system is proposed to perform MASO according to a preoperative plan and under guidance of a surgeon. A task decomposition methodology is used to divide the collaborative surgical procedure into three subtasks: (1) positional control and (2) orientation control, both led by the robot for precise alignment; and (3) force-control, managed by surgeon to ensure safety. Additionally, to achieve patient tracking without the need for a skull clamp, an optical tracking system (OTS) is utilized. Movement of the patient mandibular is measured with an optical-based tracker mounted on a dental occlusal splint. A registration method and Robot-OTS calibration method are introduced to achieve reliable navigation within our framework. The experiments of drilling were conducted on the realistic phantom model, which demonstrated that the average error between the planned and actual drilling points is 1.85mm.
X-RAFT: Cross-Modal Non-Rigid Registration of Blue and White Light Neurosurgical Hyperspectral Images
Jul 10, 2025Abstract:Integration of hyperspectral imaging into fluorescence-guided neurosurgery has the potential to improve surgical decision making by providing quantitative fluorescence measurements in real-time. Quantitative fluorescence requires paired spectral data in fluorescence (blue light) and reflectance (white light) mode. Blue and white image acquisition needs to be performed sequentially in a potentially dynamic surgical environment. A key component to the fluorescence quantification process is therefore the ability to find dense cross-modal image correspondences between two hyperspectral images taken under these drastically different lighting conditions. We address this challenge with the introduction of X-RAFT, a Recurrent All-Pairs Field Transforms (RAFT) optical flow model modified for cross-modal inputs. We propose using distinct image encoders for each modality pair, and fine-tune these in a self-supervised manner using flow-cycle-consistency on our neurosurgical hyperspectral data. We show an error reduction of 36.6% across our evaluation metrics when comparing to a naive baseline and 27.83% reduction compared to an existing cross-modal optical flow method (CrossRAFT). Our code and models will be made publicly available after the review process.
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
Average Calibration Losses for Reliable Uncertainty in Medical Image Segmentation
Jun 04, 2025Abstract:Deep neural networks for medical image segmentation are often overconfident, compromising both reliability and clinical utility. In this work, we propose differentiable formulations of marginal L1 Average Calibration Error (mL1-ACE) as an auxiliary loss that can be computed on a per-image basis. We compare both hard- and soft-binning approaches to directly improve pixel-wise calibration. Our experiments on four datasets (ACDC, AMOS, KiTS, BraTS) demonstrate that incorporating mL1-ACE significantly reduces calibration errors, particularly Average Calibration Error (ACE) and Maximum Calibration Error (MCE), while largely maintaining high Dice Similarity Coefficients (DSCs). We find that the soft-binned variant yields the greatest improvements in calibration, over the Dice plus cross-entropy loss baseline, but often compromises segmentation performance, with hard-binned mL1-ACE maintaining segmentation performance, albeit with weaker calibration improvement. To gain further insight into calibration performance and its variability across an imaging dataset, we introduce dataset reliability histograms, an aggregation of per-image reliability diagrams. The resulting analysis highlights improved alignment between predicted confidences and true accuracies. Overall, our approach not only enhances the trustworthiness of segmentation predictions but also shows potential for safer integration of deep learning methods into clinical workflows. We share our code here: https://github.com/cai4cai/Average-Calibration-Losses
A generalisable head MRI defacing pipeline: Evaluation on 2,566 meningioma scans
May 19, 2025Abstract:Reliable MRI defacing techniques to safeguard patient privacy while preserving brain anatomy are critical for research collaboration. Existing methods often struggle with incomplete defacing or degradation of brain tissue regions. We present a robust, generalisable defacing pipeline for high-resolution MRI that integrates atlas-based registration with brain masking. Our method was evaluated on 2,566 heterogeneous clinical scans for meningioma and achieved a 99.92 per cent success rate (2,564/2,566) upon visual inspection. Excellent anatomical preservation is demonstrated with a Dice similarity coefficient of 0.9975 plus or minus 0.0023 between brain masks automatically extracted from the original and defaced volumes. Source code is available at https://github.com/cai4cai/defacing_pipeline.
Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS) challenge results
May 13, 2025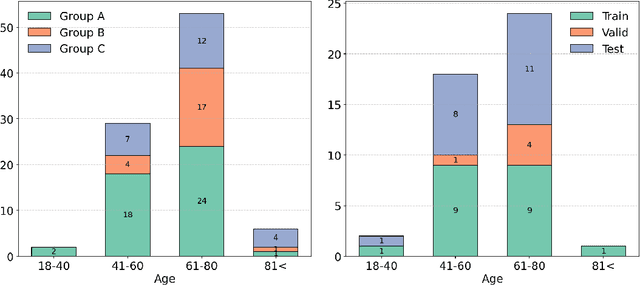
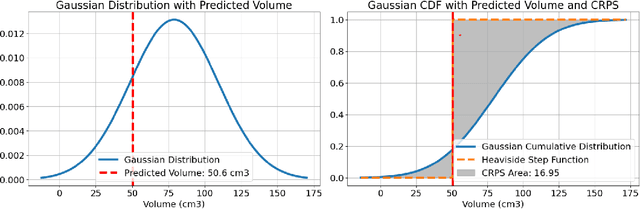

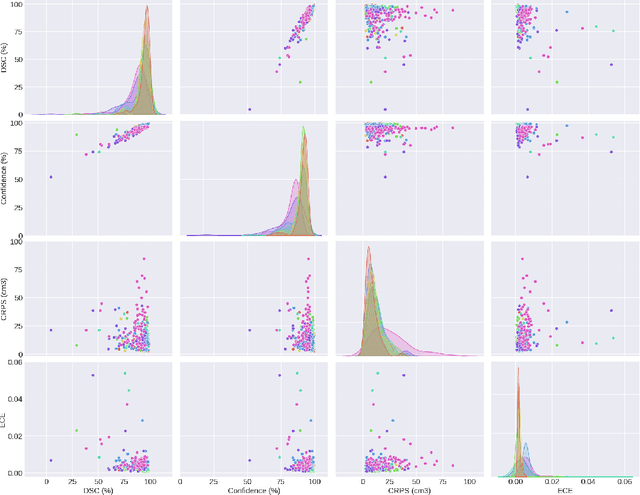
Abstract:Deep learning (DL) has become the dominant approach for medical image segmentation, yet ensuring the reliability and clinical applicability of these models requires addressing key challenges such as annotation variability, calibration, and uncertainty estimation. This is why we created the Calibration and Uncertainty for multiRater Volume Assessment in multiorgan Segmentation (CURVAS), which highlights the critical role of multiple annotators in establishing a more comprehensive ground truth, emphasizing that segmentation is inherently subjective and that leveraging inter-annotator variability is essential for robust model evaluation. Seven teams participated in the challenge, submitting a variety of DL models evaluated using metrics such as Dice Similarity Coefficient (DSC), Expected Calibration Error (ECE), and Continuous Ranked Probability Score (CRPS). By incorporating consensus and dissensus ground truth, we assess how DL models handle uncertainty and whether their confidence estimates align with true segmentation performance. Our findings reinforce the importance of well-calibrated models, as better calibration is strongly correlated with the quality of the results. Furthermore, we demonstrate that segmentation models trained on diverse datasets and enriched with pre-trained knowledge exhibit greater robustness, particularly in cases deviating from standard anatomical structures. Notably, the best-performing models achieved high DSC and well-calibrated uncertainty estimates. This work underscores the need for multi-annotator ground truth, thorough calibration assessments, and uncertainty-aware evaluations to develop trustworthy and clinically reliable DL-based medical image segmentation models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge