Navodini Wijethilake
Systematic Review of Pituitary Gland and Pituitary Adenoma Automatic Segmentation Techniques in Magnetic Resonance Imaging
Jun 24, 2025Abstract:Purpose: Accurate segmentation of both the pituitary gland and adenomas from magnetic resonance imaging (MRI) is essential for diagnosis and treatment of pituitary adenomas. This systematic review evaluates automatic segmentation methods for improving the accuracy and efficiency of MRI-based segmentation of pituitary adenomas and the gland itself. Methods: We reviewed 34 studies that employed automatic and semi-automatic segmentation methods. We extracted and synthesized data on segmentation techniques and performance metrics (such as Dice overlap scores). Results: The majority of reviewed studies utilized deep learning approaches, with U-Net-based models being the most prevalent. Automatic methods yielded Dice scores of 0.19--89.00\% for pituitary gland and 4.60--96.41\% for adenoma segmentation. Semi-automatic methods reported 80.00--92.10\% for pituitary gland and 75.90--88.36\% for adenoma segmentation. Conclusion: Most studies did not report important metrics such as MR field strength, age and adenoma size. Automated segmentation techniques such as U-Net-based models show promise, especially for adenoma segmentation, but further improvements are needed to achieve consistently good performance in small structures like the normal pituitary gland. Continued innovation and larger, diverse datasets are likely critical to enhancing clinical applicability.
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
A Clinical Guideline Driven Automated Linear Feature Extraction for Vestibular Schwannoma
Oct 30, 2023Abstract:Vestibular Schwannoma is a benign brain tumour that grows from one of the balance nerves. Patients may be treated by surgery, radiosurgery or with a conservative "wait-and-scan" strategy. Clinicians typically use manually extracted linear measurements to aid clinical decision making. This work aims to automate and improve this process by using deep learning based segmentation to extract relevant clinical features through computational algorithms. To the best of our knowledge, our study is the first to propose an automated approach to replicate local clinical guidelines. Our deep learning based segmentation provided Dice-scores of 0.8124 +- 0.2343 and 0.8969 +- 0.0521 for extrameatal and whole tumour regions respectively for T2 weighted MRI, whereas 0.8222 +- 0.2108 and 0.9049 +- 0.0646 were obtained for T1 weighted MRI. We propose a novel algorithm to choose and extract the most appropriate maximum linear measurement from the segmented regions based on the size of the extrameatal portion of the tumour. Using this tool, clinicians will be provided with a visual guide and related metrics relating to tumour progression that will function as a clinical decision aid. In this study, we utilize 187 scans obtained from 50 patients referred to a tertiary specialist neurosurgical service in the United Kingdom. The measurements extracted manually by an expert neuroradiologist indicated a significant correlation with the automated measurements (p < 0.0001).
DEEP$^2$: Deep Learning Powered De-scattering with Excitation Patterning
Oct 19, 2022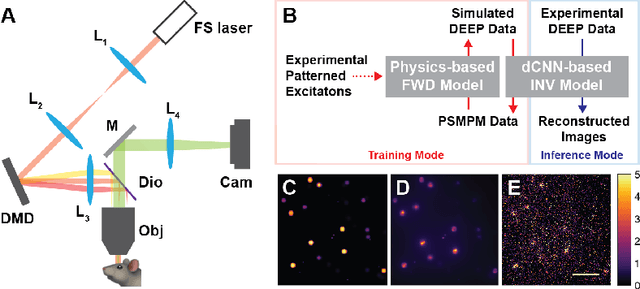
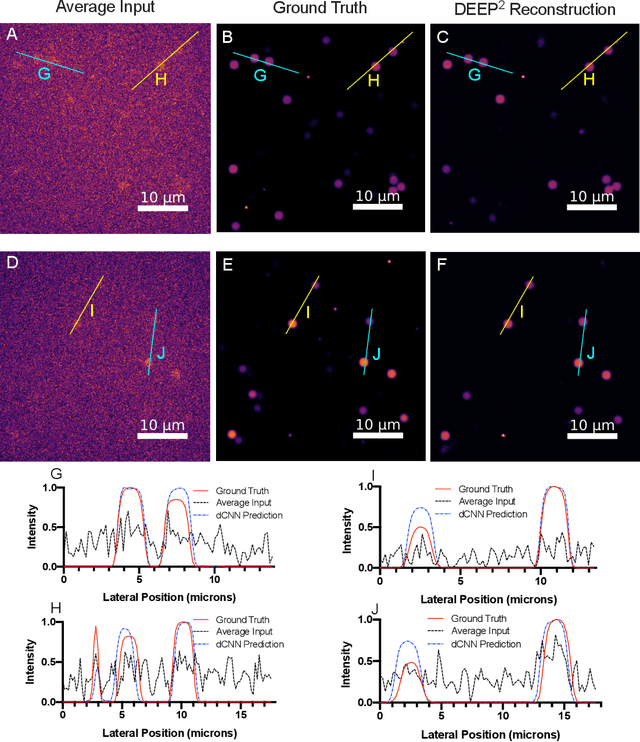
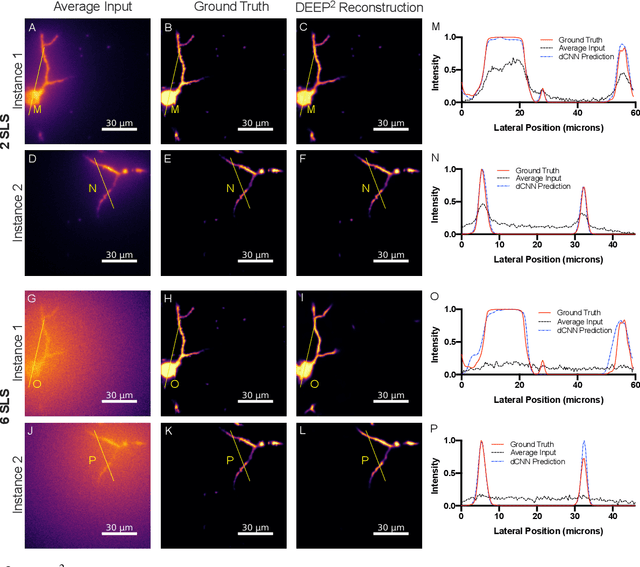
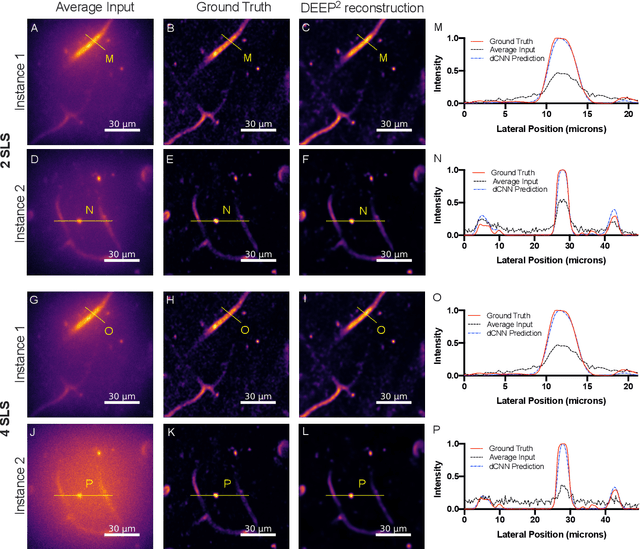
Abstract:Limited throughput is a key challenge in in-vivo deep-tissue imaging using nonlinear optical microscopy. Point scanning multiphoton microscopy, the current gold standard, is slow especially compared to the wide-field imaging modalities used for optically cleared or thin specimens. We recently introduced 'De-scattering with Excitation Patterning or DEEP', as a widefield alternative to point-scanning geometries. Using patterned multiphoton excitation, DEEP encodes spatial information inside tissue before scattering. However, to de-scatter at typical depths, hundreds of such patterned excitations are needed. In this work, we present DEEP$^2$, a deep learning based model, that can de-scatter images from just tens of patterned excitations instead of hundreds. Consequently, we improve DEEP's throughput by almost an order of magnitude. We demonstrate our method in multiple numerical and physical experiments including in-vivo cortical vasculature imaging up to four scattering lengths deep, in alive mice.
Boundary Distance Loss for Intra-/Extra-meatal Segmentation of Vestibular Schwannoma
Aug 09, 2022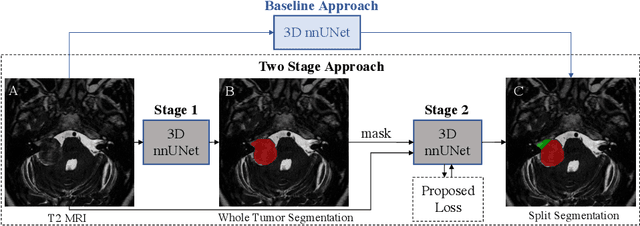
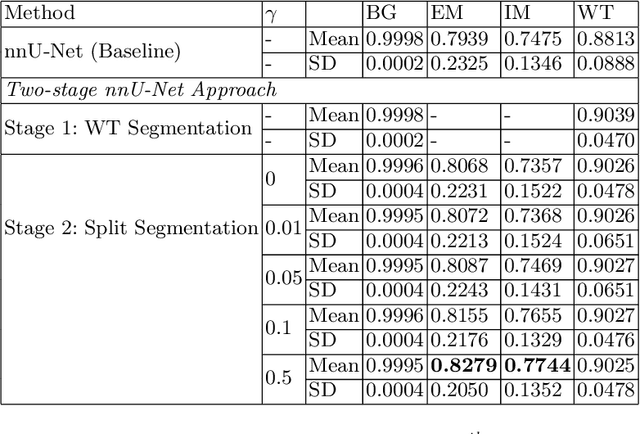
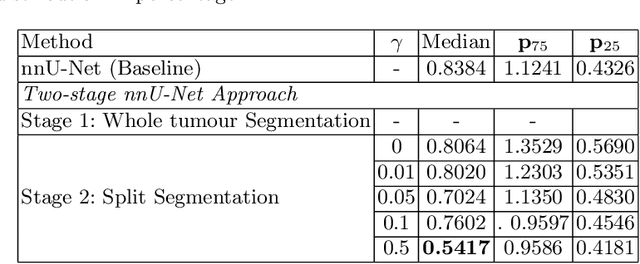
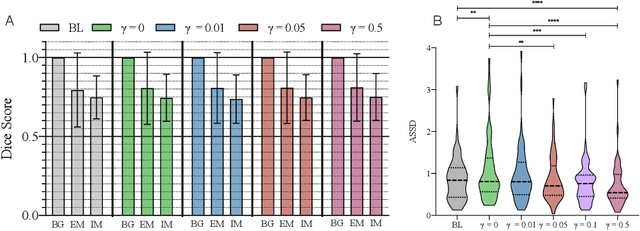
Abstract:Vestibular Schwannoma (VS) typically grows from the inner ear to the brain. It can be separated into two regions, intrameatal and extrameatal respectively corresponding to being inside or outside the inner ear canal. The growth of the extrameatal regions is a key factor that determines the disease management followed by the clinicians. In this work, a VS segmentation approach with subdivision into intra-/extra-meatal parts is presented. We annotated a dataset consisting of 227 T2 MRI instances, acquired longitudinally on 137 patients, excluding post-operative instances. We propose a staged approach, with the first stage performing the whole tumour segmentation and the second stage performing the intra-/extra-meatal segmentation using the T2 MRI along with the mask obtained from the first stage. To improve on the accuracy of the predicted meatal boundary, we introduce a task-specific loss which we call Boundary Distance Loss. The performance is evaluated in contrast to the direct intrameatal extrameatal segmentation task performance, i.e. the Baseline. Our proposed method, with the two-stage approach and the Boundary Distance Loss, achieved a Dice score of 0.8279+-0.2050 and 0.7744+-0.1352 for extrameatal and intrameatal regions respectively, significantly improving over the Baseline, which gave Dice score of 0.7939+-0.2325 and 0.7475+-0.1346 for the extrameatal and intrameatal regions respectively.
Brain Tumor Segmentation and Survival Prediction using 3D Attention UNet
Apr 02, 2021



Abstract:In this work, we develop an attention convolutional neural network (CNN) to segment brain tumors from Magnetic Resonance Images (MRI). Further, we predict the survival rate using various machine learning methods. We adopt a 3D UNet architecture and integrate channel and spatial attention with the decoder network to perform segmentation. For survival prediction, we extract some novel radiomic features based on geometry, location, the shape of the segmented tumor and combine them with clinical information to estimate the survival duration for each patient. We also perform extensive experiments to show the effect of each feature for overall survival (OS) prediction. The experimental results infer that radiomic features such as histogram, location, and shape of the necrosis region and clinical features like age are the most critical parameters to estimate the OS.
Glioblastoma Multiforme Prognosis: MRI Missing Modality Generation, Segmentation and Radiogenomic Survival Prediction
Mar 17, 2021



Abstract:The accurate prognosis of Glioblastoma Multiforme (GBM) plays an essential role in planning correlated surgeries and treatments. The conventional models of survival prediction rely on radiomic features using magnetic resonance imaging (MRI). In this paper, we propose a radiogenomic overall survival (OS) prediction approach by incorporating gene expression data with radiomic features such as shape, geometry, and clinical information. We exploit TCGA (The Cancer Genomic Atlas) dataset and synthesize the missing MRI modalities using a fully convolutional network (FCN) in a conditional Generative Adversarial Network (cGAN). Meanwhile, the same FCN architecture enables the tumor segmentation from the available and the synthesized MRI modalities. The proposed FCN architecture comprises octave convolution (OctConv) and a novel decoder, with skip connections in spatial and channel squeeze & excitation (skip-scSE) block. The OctConv can process low and high-frequency features individually and improve model efficiency by reducing channel-wise redundancy. Skip-scSE applies spatial and channel-wise excitation to signify the essential features and reduces the sparsity in deeper layers learning parameters using skip connections. The proposed approaches are evaluated by comparative experiments with state-of-the-art models in synthesis, segmentation, and overall survival (OS) prediction. We observe that adding missing MRI modality improves the segmentation prediction, and expression levels of gene markers have a high contribution in the GBM prognosis prediction, and fused radiogenomic features boost the OS estimation.
A Thickness Sensitive Vessel Extraction Framework for Retinal and Conjunctival Vascular Tortuosity Analysis
Jan 02, 2021
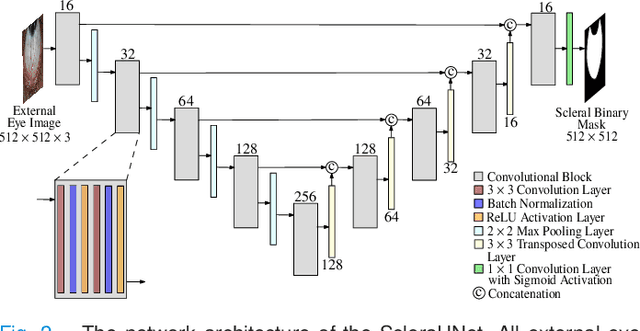
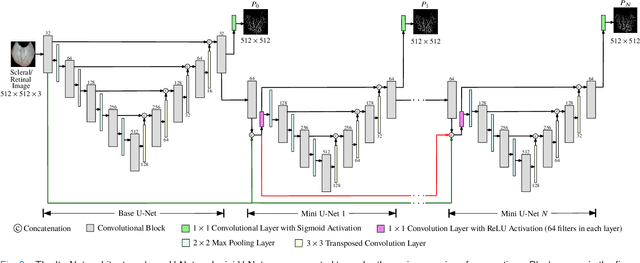

Abstract:Systemic diseases such as diabetes, hypertension, atherosclerosis are among the leading causes of annual human mortality rate. It is suggested that retinal and conjunctival vascular tortuosity is a potential biomarker for such systemic diseases. Most importantly, it is observed that the tortuosity depends on the thickness of these vessels. Therefore, selective calculation of tortuosity within specific vessel thicknesses is required depending on the disease being analysed. In this paper, we propose a thickness sensitive vessel extraction framework that is primarily applicable for studies related to retinal and conjunctival vascular tortuosity. The framework uses a Convolutional Neural Network based on the IterNet architecture to obtain probability maps of the entire vasculature. They are then processed by a multi-scale vessel enhancement technique that exploits both fine and coarse structural vascular details of these probability maps in order to extract vessels of specified thicknesses. We evaluated the proposed framework on four datasets including DRIVE and SBVPI, and obtained Matthew's Correlation Coefficient values greater than 0.71 for all the datasets. In addition, the proposed framework was utilized to determine the association of diabetes with retinal and conjunctival vascular tortuosity. We observed that retinal vascular tortuosity (Eccentricity based Tortuosity Index) of the diabetic group was significantly higher (p < .05) than that of the non-diabetic group and that conjunctival vascular tortuosity (Total Curvature normalized by Arc Length) of diabetic group was significantly lower (p < .05) than that of the non-diabetic group. These observations were in agreement with the literature, strengthening the suitability of the proposed framework.
Survival prediction and risk estimation of Glioma patients using mRNA expressions
Nov 02, 2020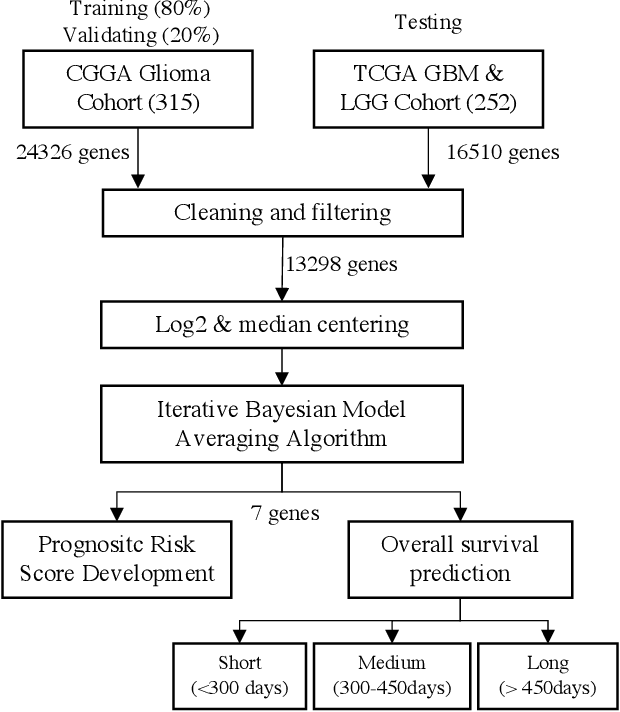
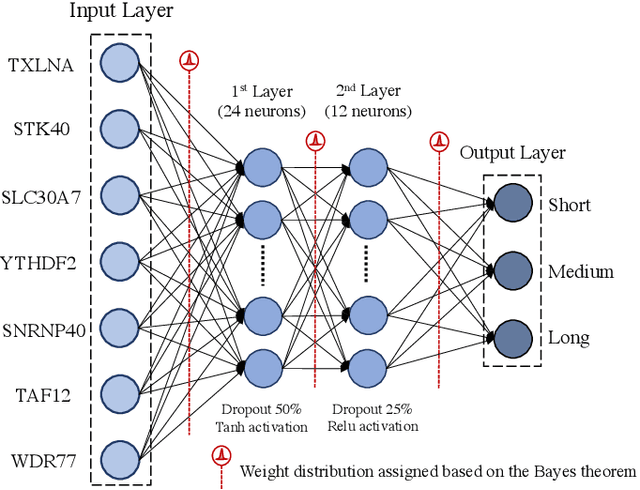
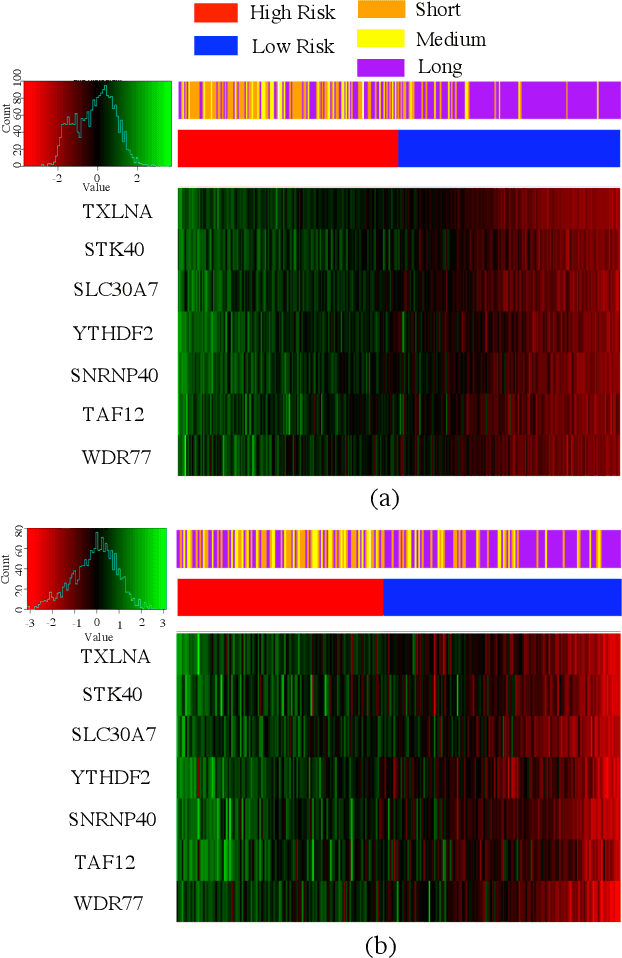
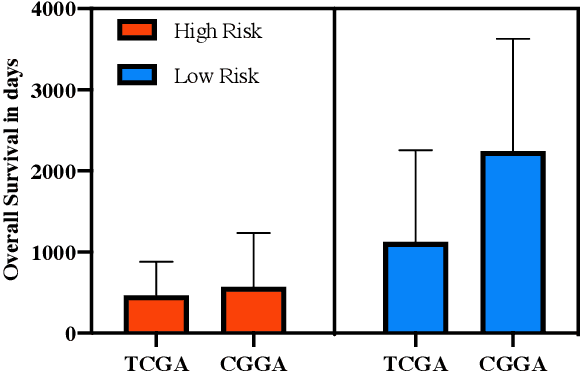
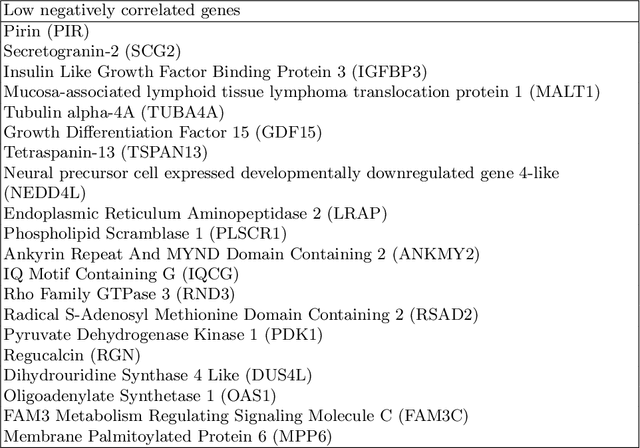
Abstract:Gliomas are lethal type of central nervous system tumors with a poor prognosis. Recently, with the advancements in the micro-array technologies thousands of gene expression related data of glioma patients are acquired, leading for salient analysis in many aspects. Thus, genomics are been emerged into the field of prognosis analysis. In this work, we identify survival related 7 gene signature and explore two approaches for survival prediction and risk estimation. For survival prediction, we propose a novel probabilistic programming based approach, which outperforms the existing traditional machine learning algorithms. An average 4 fold accuracy of 74% is obtained with the proposed algorithm. Further, we construct a prognostic risk model for risk estimation of glioma patients. This model reflects the survival of glioma patients, with high risk for low survival patients.
Radiogenomics of Glioblastoma: Identification of Radiomics associated with Molecular Subtypes
Oct 27, 2020
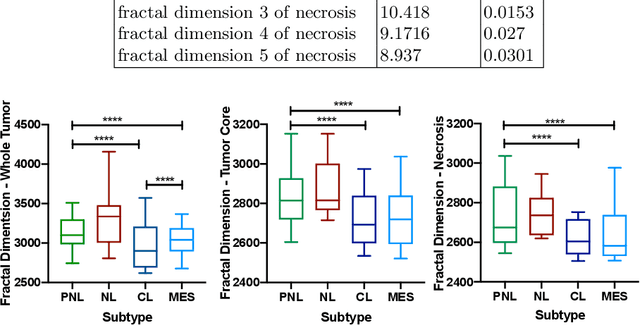
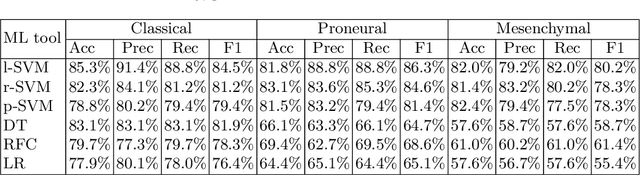
Abstract:Glioblastoma is the most malignant type of central nervous system tumor with GBM subtypes cleaved based on molecular level gene alterations. These alterations are also happened to affect the histology. Thus, it can cause visible changes in images, such as enhancement and edema development. In this study, we extract intensity, volume, and texture features from the tumor subregions to identify the correlations with gene expression features and overall survival. Consequently, we utilize the radiomics to find associations with the subtypes of glioblastoma. Accordingly, the fractal dimensions of the whole tumor, tumor core, and necrosis regions show a significant difference between the Proneural, Classical and Mesenchymal subtypes. Additionally, the subtypes of GBM are predicted with an average accuracy of 79% utilizing radiomics and accuracy over 90% utilizing gene expression profiles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge