Muhammad Asad
Confidence-aware Monocular Depth Estimation for Minimally Invasive Surgery
Mar 03, 2026Abstract:Purpose: Monocular depth estimation (MDE) is vital for scene understanding in minimally invasive surgery (MIS). However, endoscopic video sequences are often contaminated by smoke, specular reflections, blur, and occlusions, limiting the accuracy of MDE models. In addition, current MDE models do not output depth confidence, which could be a valuable tool for improving their clinical reliability. Methods: We propose a novel confidence-aware MDE framework featuring three significant contributions: (i) Calibrated confidence targets: an ensemble of fine-tuned stereo matching models is used to capture disparity variance into pixel-wise confidence probabilities; (ii) Confidence-aware loss: Baseline MDE models are optimized with confidence-aware loss functions, utilizing pixel-wise confidence probabilities such that reliable pixels dominate training; and (iii) Inference-time confidence: a confidence estimation head is proposed with two convolution layers to predict per-pixel confidence at inference, enabling assessment of depth reliability. Results: Comprehensive experimental validation across internal and public datasets demonstrates that our framework improves depth estimation accuracy and can robustly quantify the prediction's confidence. On the internal clinical endoscopic dataset (StereoKP), we improve dense depth estimation accuracy by ~8% as compared to the baseline model. Conclusion: Our confidence-aware framework enables improved accuracy of MDE models in MIS, addressing challenges posed by noise and artifacts in pre-clinical and clinical data, and allows MDE models to provide confidence maps that may be used to improve their reliability for clinical applications.
HanDrawer: Leveraging Spatial Information to Render Realistic Hands Using a Conditional Diffusion Model in Single Stage
Mar 03, 2025



Abstract:Although diffusion methods excel in text-to-image generation, generating accurate hand gestures remains a major challenge, resulting in severe artifacts, such as incorrect number of fingers or unnatural gestures. To enable the diffusion model to learn spatial information to improve the quality of the hands generated, we propose HanDrawer, a module to condition the hand generation process. Specifically, we apply graph convolutional layers to extract the endogenous spatial structure and physical constraints implicit in MANO hand mesh vertices. We then align and fuse these spatial features with other modalities via cross-attention. The spatially fused features are used to guide a single stage diffusion model denoising process for high quality generation of the hand region. To improve the accuracy of spatial feature fusion, we propose a Position-Preserving Zero Padding (PPZP) fusion strategy, which ensures that the features extracted by HanDrawer are fused into the region of interest in the relevant layers of the diffusion model. HanDrawer learns the entire image features while paying special attention to the hand region thanks to an additional hand reconstruction loss combined with the denoising loss. To accurately train and evaluate our approach, we perform careful cleansing and relabeling of the widely used HaGRID hand gesture dataset and obtain high quality multimodal data. Quantitative and qualitative analyses demonstrate the state-of-the-art performance of our method on the HaGRID dataset through multiple evaluation metrics. Source code and our enhanced dataset will be released publicly if the paper is accepted.
Compositional Segmentation of Cardiac Images Leveraging Metadata
Oct 30, 2024Abstract:Cardiac image segmentation is essential for automated cardiac function assessment and monitoring of changes in cardiac structures over time. Inspired by coarse-to-fine approaches in image analysis, we propose a novel multitask compositional segmentation approach that can simultaneously localize the heart in a cardiac image and perform part-based segmentation of different regions of interest. We demonstrate that this compositional approach achieves better results than direct segmentation of the anatomies. Further, we propose a novel Cross-Modal Feature Integration (CMFI) module to leverage the metadata related to cardiac imaging collected during image acquisition. We perform experiments on two different modalities, MRI and ultrasound, using public datasets, Multi-disease, Multi-View, and Multi-Centre (M&Ms-2) and Multi-structure Ultrasound Segmentation (CAMUS) data, to showcase the efficiency of the proposed compositional segmentation method and Cross-Modal Feature Integration module incorporating metadata within the proposed compositional segmentation network. The source code is available: https://github.com/kabbas570/CompSeg-MetaData.
Adaptive Multi-Modal Control of Digital Human Hand Synthesis Using a Region-Aware Cycle Loss
Sep 13, 2024



Abstract:Diffusion models have shown their remarkable ability to synthesize images, including the generation of humans in specific poses. However, current models face challenges in adequately expressing conditional control for detailed hand pose generation, leading to significant distortion in the hand regions. To tackle this problem, we first curate the How2Sign dataset to provide richer and more accurate hand pose annotations. In addition, we introduce adaptive, multi-modal fusion to integrate characters' physical features expressed in different modalities such as skeleton, depth, and surface normal. Furthermore, we propose a novel Region-Aware Cycle Loss (RACL) that enables the diffusion model training to focus on improving the hand region, resulting in improved quality of generated hand gestures. More specifically, the proposed RACL computes a weighted keypoint distance between the full-body pose keypoints from the generated image and the ground truth, to generate higher-quality hand poses while balancing overall pose accuracy. Moreover, we use two hand region metrics, named hand-PSNR and hand-Distance for hand pose generation evaluations. Our experimental evaluations demonstrate the effectiveness of our proposed approach in improving the quality of digital human pose generation using diffusion models, especially the quality of the hand region. The source code is available at https://github.com/fuqifan/Region-Aware-Cycle-Loss.
Convolution and Attention-Free Mamba-based Cardiac Image Segmentation
Jun 09, 2024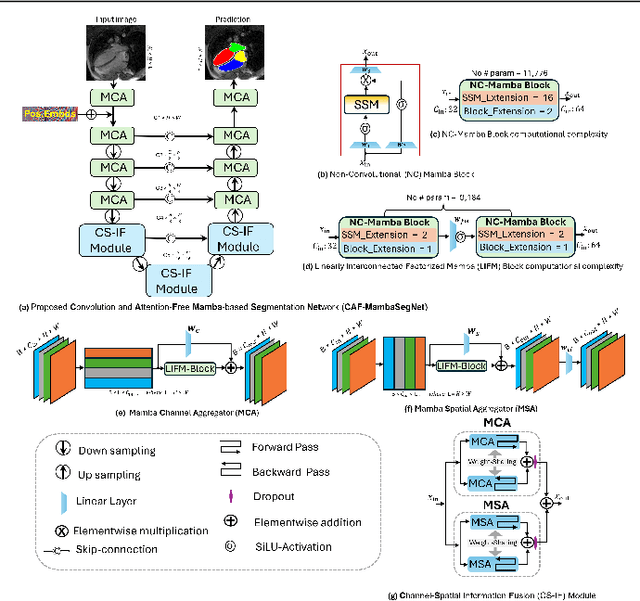
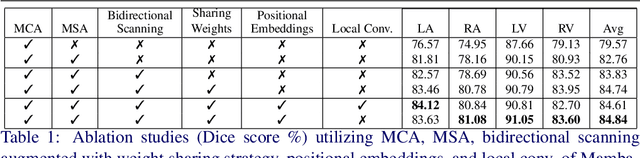
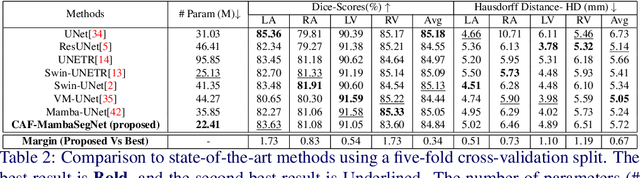
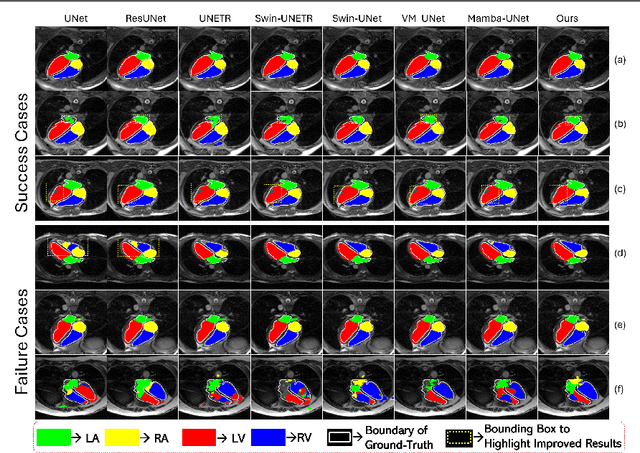
Abstract:Convolutional Neural Networks (CNNs) and Transformer-based self-attention models have become standard for medical image segmentation. This paper demonstrates that convolution and self-attention, while widely used, are not the only effective methods for segmentation. Breaking with convention, we present a Convolution and self-Attention Free Mamba-based semantic Segmentation Network named CAF-MambaSegNet. Specifically, we design a Mamba-based Channel Aggregator and Spatial Aggregator, which are applied independently in each encoder-decoder stage. The Channel Aggregator extracts information across different channels, and the Spatial Aggregator learns features across different spatial locations. We also propose a Linearly Interconnected Factorized Mamba (LIFM) Block to reduce the computational complexity of a Mamba and to enhance its decision function by introducing a non-linearity between two factorized Mamba blocks. Our goal is not to outperform state-of-the-art results but to show how this innovative, convolution and self-attention-free method can inspire further research beyond well-established CNNs and Transformers, achieving linear complexity and reducing the number of parameters. Source code and pre-trained models will be publicly available.
Multi-view Cardiac Image Segmentation via Trans-Dimensional Priors
Apr 25, 2024



Abstract:We propose a novel multi-stage trans-dimensional architecture for multi-view cardiac image segmentation. Our method exploits the relationship between long-axis (2D) and short-axis (3D) magnetic resonance (MR) images to perform a sequential 3D-to-2D-to-3D segmentation, segmenting the long-axis and short-axis images. In the first stage, 3D segmentation is performed using the short-axis image, and the prediction is transformed to the long-axis view and used as a segmentation prior in the next stage. In the second step, the heart region is localized and cropped around the segmentation prior using a Heart Localization and Cropping (HLC) module, focusing the subsequent model on the heart region of the image, where a 2D segmentation is performed. Similarly, we transform the long-axis prediction to the short-axis view, localize and crop the heart region and again perform a 3D segmentation to refine the initial short-axis segmentation. We evaluate our proposed method on the Multi-Disease, Multi-View & Multi-Center Right Ventricular Segmentation in Cardiac MRI (M&Ms-2) dataset, where our method outperforms state-of-the-art methods in segmenting cardiac regions of interest in both short-axis and long-axis images. The pre-trained models, source code, and implementation details will be publicly available.
Beyond the Known: Adversarial Autoencoders in Novelty Detection
Apr 06, 2024Abstract:In novelty detection, the goal is to decide if a new data point should be categorized as an inlier or an outlier, given a training dataset that primarily captures the inlier distribution. Recent approaches typically use deep encoder and decoder network frameworks to derive a reconstruction error, and employ this error either to determine a novelty score, or as the basis for a one-class classifier. In this research, we use a similar framework but with a lightweight deep network, and we adopt a probabilistic score with reconstruction error. Our methodology calculates the probability of whether the sample comes from the inlier distribution or not. This work makes two key contributions. The first is that we compute the novelty probability by linearizing the manifold that holds the structure of the inlier distribution. This allows us to interpret how the probability is distributed and can be determined in relation to the local coordinates of the manifold tangent space. The second contribution is that we improve the training protocol for the network. Our results indicate that our approach is effective at learning the target class, and it outperforms recent state-of-the-art methods on several benchmark datasets.
Crop and Couple: cardiac image segmentation using interlinked specialist networks
Feb 14, 2024Abstract:Diagnosis of cardiovascular disease using automated methods often relies on the critical task of cardiac image segmentation. We propose a novel strategy that performs segmentation using specialist networks that focus on a single anatomy (left ventricle, right ventricle, or myocardium). Given an input long-axis cardiac MR image, our method performs a ternary segmentation in the first stage to identify these anatomical regions, followed by cropping the original image to focus subsequent processing on the anatomical regions. The specialist networks are coupled through an attention mechanism that performs cross-attention to interlink features from different anatomies, serving as a soft relative shape prior. Central to our approach is an additive attention block (E-2A block), which is used throughout our architecture thanks to its efficiency.
DEEPBEAS3D: Deep Learning and B-Spline Explicit Active Surfaces
Sep 05, 2023Abstract:Deep learning-based automatic segmentation methods have become state-of-the-art. However, they are often not robust enough for direct clinical application, as domain shifts between training and testing data affect their performance. Failure in automatic segmentation can cause sub-optimal results that require correction. To address these problems, we propose a novel 3D extension of an interactive segmentation framework that represents a segmentation from a convolutional neural network (CNN) as a B-spline explicit active surface (BEAS). BEAS ensures segmentations are smooth in 3D space, increasing anatomical plausibility, while allowing the user to precisely edit the 3D surface. We apply this framework to the task of 3D segmentation of the anal sphincter complex (AS) from transperineal ultrasound (TPUS) images, and compare it to the clinical tool used in the pelvic floor disorder clinic (4D View VOCAL, GE Healthcare; Zipf, Austria). Experimental results show that: 1) the proposed framework gives the user explicit control of the surface contour; 2) the perceived workload calculated via the NASA-TLX index was reduced by 30% compared to VOCAL; and 3) it required 7 0% (170 seconds) less user time than VOCAL (p< 0.00001)
DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images
May 18, 2023Abstract:Automatic segmentation of medical images is a key step for diagnostic and interventional tasks. However, achieving this requires large amounts of annotated volumes, which can be tedious and time-consuming task for expert annotators. In this paper, we introduce DeepEdit, a deep learning-based method for volumetric medical image annotation, that allows automatic and semi-automatic segmentation, and click-based refinement. DeepEdit combines the power of two methods: a non-interactive (i.e. automatic segmentation using nnU-Net, UNET or UNETR) and an interactive segmentation method (i.e. DeepGrow), into a single deep learning model. It allows easy integration of uncertainty-based ranking strategies (i.e. aleatoric and epistemic uncertainty computation) and active learning. We propose and implement a method for training DeepEdit by using standard training combined with user interaction simulation. Once trained, DeepEdit allows clinicians to quickly segment their datasets by using the algorithm in auto segmentation mode or by providing clicks via a user interface (i.e. 3D Slicer, OHIF). We show the value of DeepEdit through evaluation on the PROSTATEx dataset for prostate/prostatic lesions and the Multi-Atlas Labeling Beyond the Cranial Vault (BTCV) dataset for abdominal CT segmentation, using state-of-the-art network architectures as baseline for comparison. DeepEdit could reduce the time and effort annotating 3D medical images compared to DeepGrow alone. Source code is available at https://github.com/Project-MONAI/MONAILabel
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge