Indrajeet Mandal
Probing the limitations of multimodal language models for chemistry and materials research
Nov 25, 2024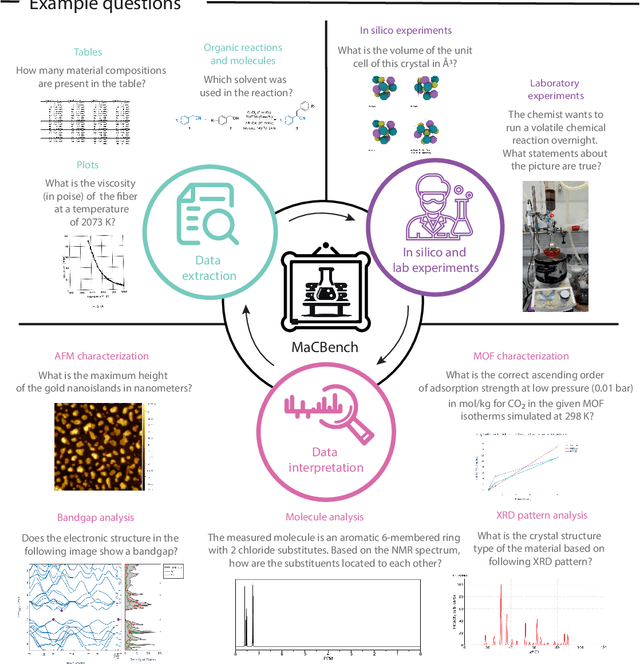
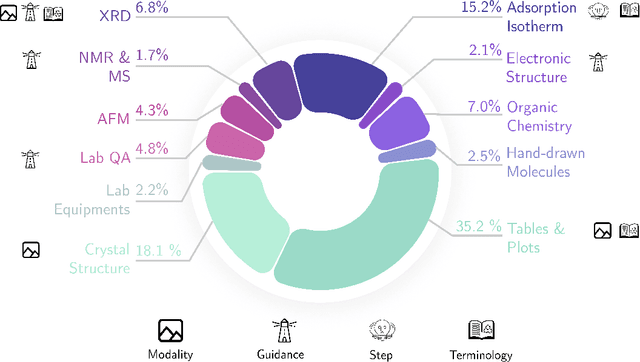
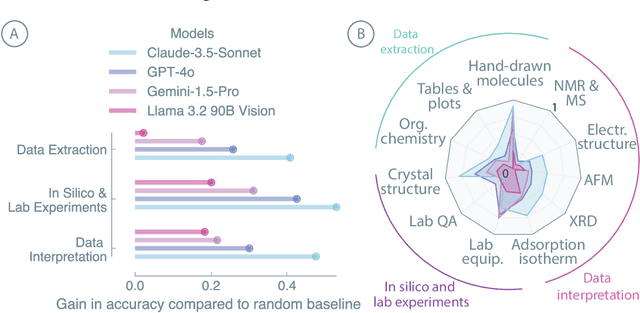
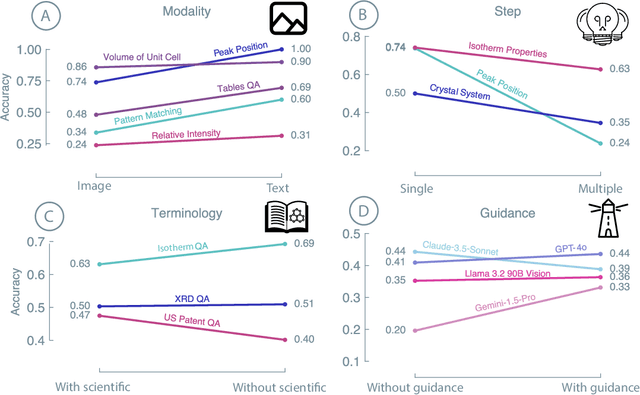
Abstract:Recent advancements in artificial intelligence have sparked interest in scientific assistants that could support researchers across the full spectrum of scientific workflows, from literature review to experimental design and data analysis. A key capability for such systems is the ability to process and reason about scientific information in both visual and textual forms - from interpreting spectroscopic data to understanding laboratory setups. Here, we introduce MaCBench, a comprehensive benchmark for evaluating how vision-language models handle real-world chemistry and materials science tasks across three core aspects: data extraction, experimental understanding, and results interpretation. Through a systematic evaluation of leading models, we find that while these systems show promising capabilities in basic perception tasks - achieving near-perfect performance in equipment identification and standardized data extraction - they exhibit fundamental limitations in spatial reasoning, cross-modal information synthesis, and multi-step logical inference. Our insights have important implications beyond chemistry and materials science, suggesting that developing reliable multimodal AI scientific assistants may require advances in curating suitable training data and approaches to training those models.
Adaptive Multi-scale Online Likelihood Network for AI-assisted Interactive Segmentation
Mar 23, 2023Abstract:Existing interactive segmentation methods leverage automatic segmentation and user interactions for label refinement, significantly reducing the annotation workload compared to manual annotation. However, these methods lack quick adaptability to ambiguous and noisy data, which is a challenge in CT volumes containing lung lesions from COVID-19 patients. In this work, we propose an adaptive multi-scale online likelihood network (MONet) that adaptively learns in a data-efficient online setting from both an initial automatic segmentation and user interactions providing corrections. We achieve adaptive learning by proposing an adaptive loss that extends the influence of user-provided interaction to neighboring regions with similar features. In addition, we propose a data-efficient probability-guided pruning method that discards uncertain and redundant labels in the initial segmentation to enable efficient online training and inference. Our proposed method was evaluated by an expert in a blinded comparative study on COVID-19 lung lesion annotation task in CT. Our approach achieved 5.86% higher Dice score with 24.67% less perceived NASA-TLX workload score than the state-of-the-art. Source code is available at: https://github.com/masadcv/MONet-MONAILabel
VertXNet: An Ensemble Method for Vertebrae Segmentation and Identification of Spinal X-Ray
Feb 07, 2023Abstract:Reliable vertebrae annotations are key to perform analysis of spinal X-ray images. However, obtaining annotation of vertebrae from those images is usually carried out manually due to its complexity (i.e. small structures with varying shape), making it a costly and tedious process. To accelerate this process, we proposed an ensemble pipeline, VertXNet, that combines two state-of-the-art (SOTA) segmentation models (respectively U-Net and Mask R-CNN) to automatically segment and label vertebrae in X-ray spinal images. Moreover, VertXNet introduces a rule-based approach that allows to robustly infer vertebrae labels (by locating the 'reference' vertebrae which are easier to segment than others) for a given spinal X-ray image. We evaluated the proposed pipeline on three spinal X-ray datasets (two internal and one publicly available), and compared against vertebrae annotated by radiologists. Our experimental results have shown that the proposed pipeline outperformed two SOTA segmentation models on our test dataset (MEASURE 1) with a mean Dice of 0.90, vs. a mean Dice of 0.73 for Mask R-CNN and 0.72 for U-Net. To further evaluate the generalization ability of VertXNet, the pre-trained pipeline was directly tested on two additional datasets (PREVENT and NHANES II) and consistent performance was observed with a mean Dice of 0.89 and 0.88, respectively. Overall, VertXNet demonstrated significantly improved performance for vertebra segmentation and labeling for spinal X-ray imaging, and evaluation on both in-house clinical trial data and publicly available data further proved its generalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge