Yuanhan Mo
Deformation-Recovery Diffusion Model (DRDM): Instance Deformation for Image Manipulation and Synthesis
Jul 10, 2024Abstract:In medical imaging, the diffusion models have shown great potential in synthetic image generation tasks. However, these models often struggle with the interpretable connections between the generated and existing images and could create illusions. To address these challenges, our research proposes a novel diffusion-based generative model based on deformation diffusion and recovery. This model, named Deformation-Recovery Diffusion Model (DRDM), diverges from traditional score/intensity and latent feature-based approaches, emphasizing morphological changes through deformation fields rather than direct image synthesis. This is achieved by introducing a topological-preserving deformation field generation method, which randomly samples and integrates a set of multi-scale Deformation Vector Fields (DVF). DRDM is trained to learn to recover unreasonable deformation components, thereby restoring each randomly deformed image to a realistic distribution. These innovations facilitate the generation of diverse and anatomically plausible deformations, enhancing data augmentation and synthesis for further analysis in downstream tasks, such as few-shot learning and image registration. Experimental results in cardiac MRI and pulmonary CT show DRDM is capable of creating diverse, large (over 10% image size deformation scale), and high-quality (negative ratio of folding rate is lower than 1%) deformation fields. The further experimental results in downstream tasks, 2D image segmentation and 3D image registration, indicate significant improvements resulting from DRDM, showcasing the potential of our model to advance image manipulation and synthesis in medical imaging and beyond. Our implementation will be available at https://github.com/jianqingzheng/def_diff_rec.
Spinal Osteophyte Detection via Robust Patch Extraction on minimally annotated X-rays
Feb 29, 2024Abstract:The development and progression of arthritis is strongly associated with osteophytes, which are small and elusive bone growths. This paper presents one of the first efforts towards automated spinal osteophyte detection in spinal X-rays. A novel automated patch extraction process, called SegPatch, has been proposed based on deep learning-driven vertebrae segmentation and the enlargement of mask contours. A final patch classification accuracy of 84.5\% is secured, surpassing a baseline tiling-based patch generation technique by 9.5%. This demonstrates that even with limited annotations, SegPatch can deliver superior performance for detection of tiny structures such as osteophytes. The proposed approach has potential to assist clinicians in expediting the process of manually identifying osteophytes in spinal X-ray.
Multi-Task Cooperative Learning via Searching for Flat Minima
Sep 21, 2023

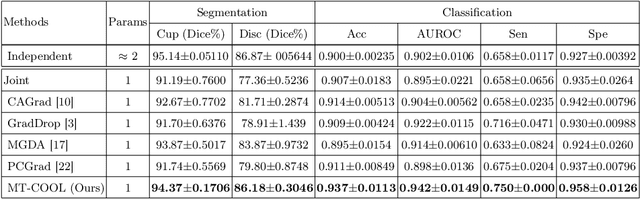

Abstract:Multi-task learning (MTL) has shown great potential in medical image analysis, improving the generalizability of the learned features and the performance in individual tasks. However, most of the work on MTL focuses on either architecture design or gradient manipulation, while in both scenarios, features are learned in a competitive manner. In this work, we propose to formulate MTL as a multi/bi-level optimization problem, and therefore force features to learn from each task in a cooperative approach. Specifically, we update the sub-model for each task alternatively taking advantage of the learned sub-models of the other tasks. To alleviate the negative transfer problem during the optimization, we search for flat minima for the current objective function with regard to features from other tasks. To demonstrate the effectiveness of the proposed approach, we validate our method on three publicly available datasets. The proposed method shows the advantage of cooperative learning, and yields promising results when compared with the state-of-the-art MTL approaches. The code will be available online.
Towards Automatic Scoring of Spinal X-ray for Ankylosing Spondylitis
Aug 08, 2023Abstract:Manually grading structural changes with the modified Stoke Ankylosing Spondylitis Spinal Score (mSASSS) on spinal X-ray imaging is costly and time-consuming due to bone shape complexity and image quality variations. In this study, we address this challenge by prototyping a 2-step auto-grading pipeline, called VertXGradeNet, to automatically predict mSASSS scores for the cervical and lumbar vertebral units (VUs) in X-ray spinal imaging. The VertXGradeNet utilizes VUs generated by our previously developed VU extraction pipeline (VertXNet) as input and predicts mSASSS based on those VUs. VertXGradeNet was evaluated on an in-house dataset of lateral cervical and lumbar X-ray images for axial spondylarthritis patients. Our results show that VertXGradeNet can predict the mSASSS score for each VU when the data is limited in quantity and imbalanced. Overall, it can achieve a balanced accuracy of 0.56 and 0.51 for 4 different mSASSS scores (i.e., a score of 0, 1, 2, 3) on two test datasets. The accuracy of the presented method shows the potential to streamline the spinal radiograph readings and therefore reduce the cost of future clinical trials.
VertXNet: An Ensemble Method for Vertebrae Segmentation and Identification of Spinal X-Ray
Feb 07, 2023Abstract:Reliable vertebrae annotations are key to perform analysis of spinal X-ray images. However, obtaining annotation of vertebrae from those images is usually carried out manually due to its complexity (i.e. small structures with varying shape), making it a costly and tedious process. To accelerate this process, we proposed an ensemble pipeline, VertXNet, that combines two state-of-the-art (SOTA) segmentation models (respectively U-Net and Mask R-CNN) to automatically segment and label vertebrae in X-ray spinal images. Moreover, VertXNet introduces a rule-based approach that allows to robustly infer vertebrae labels (by locating the 'reference' vertebrae which are easier to segment than others) for a given spinal X-ray image. We evaluated the proposed pipeline on three spinal X-ray datasets (two internal and one publicly available), and compared against vertebrae annotated by radiologists. Our experimental results have shown that the proposed pipeline outperformed two SOTA segmentation models on our test dataset (MEASURE 1) with a mean Dice of 0.90, vs. a mean Dice of 0.73 for Mask R-CNN and 0.72 for U-Net. To further evaluate the generalization ability of VertXNet, the pre-trained pipeline was directly tested on two additional datasets (PREVENT and NHANES II) and consistent performance was observed with a mean Dice of 0.89 and 0.88, respectively. Overall, VertXNet demonstrated significantly improved performance for vertebra segmentation and labeling for spinal X-ray imaging, and evaluation on both in-house clinical trial data and publicly available data further proved its generalization.
The Extreme Cardiac MRI Analysis Challenge under Respiratory Motion (CMRxMotion)
Oct 12, 2022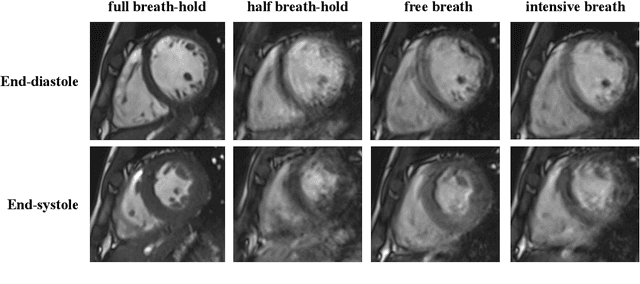
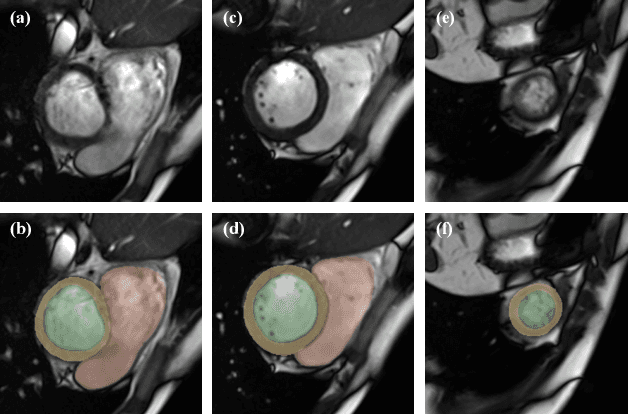

Abstract:The quality of cardiac magnetic resonance (CMR) imaging is susceptible to respiratory motion artifacts. The model robustness of automated segmentation techniques in face of real-world respiratory motion artifacts is unclear. This manuscript describes the design of extreme cardiac MRI analysis challenge under respiratory motion (CMRxMotion Challenge). The challenge aims to establish a public benchmark dataset to assess the effects of respiratory motion on image quality and examine the robustness of segmentation models. The challenge recruited 40 healthy volunteers to perform different breath-hold behaviors during one imaging visit, obtaining paired cine imaging with artifacts. Radiologists assessed the image quality and annotated the level of respiratory motion artifacts. For those images with diagnostic quality, radiologists further segmented the left ventricle, left ventricle myocardium and right ventricle. The images of training set (20 volunteers) along with the annotations are released to the challenge participants, to develop an automated image quality assessment model (Task 1) and an automated segmentation model (Task 2). The images of validation set (5 volunteers) are released to the challenge participants but the annotations are withheld for online evaluation of submitted predictions. Both the images and annotations of the test set (15 volunteers) were withheld and only used for offline evaluation of submitted containerized dockers. The image quality assessment task is quantitatively evaluated by the Cohen's kappa statistics and the segmentation task is evaluated by the Dice scores and Hausdorff distances.
VertXNet: Automatic Segmentation and Identification of Lumbar and Cervical Vertebrae from Spinal X-ray Images
Jul 12, 2022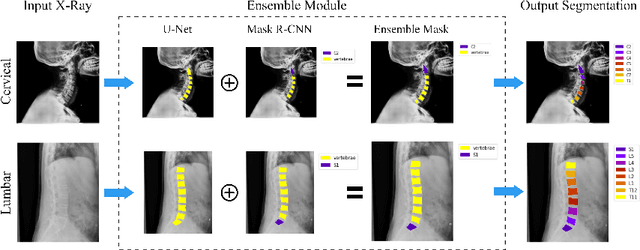
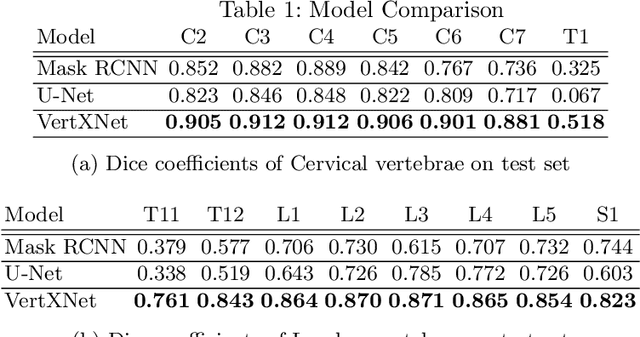
Abstract:Manual annotation of vertebrae on spinal X-ray imaging is costly and time-consuming due to bone shape complexity and image quality variations. In this study, we address this challenge by proposing an ensemble method called VertXNet, to automatically segment and label vertebrae in X-ray spinal images. VertXNet combines two state-of-the-art segmentation models, namely U-Net and Mask R-CNN to improve vertebrae segmentation. A main feature of VertXNet is to also infer vertebrae labels thanks to its Mask R-CNN component (trained to detect 'reference' vertebrae) on a given spinal X-ray image. VertXNet was evaluated on an in-house dataset of lateral cervical and lumbar X-ray imaging for ankylosing spondylitis (AS) patients. Our results show that VertXNet can accurately label spinal X-rays (mean Dice of 0.9). It can be used to circumvent the lack of annotated vertebrae without requiring human expert review. This step is crucial to investigate clinical associations by solving the lack of segmentation, a common bottleneck for most computational imaging projects.
Suggestive Annotation of Brain MR Images with Gradient-guided Sampling
Jun 02, 2022



Abstract:Machine learning has been widely adopted for medical image analysis in recent years given its promising performance in image segmentation and classification tasks. The success of machine learning, in particular supervised learning, depends on the availability of manually annotated datasets. For medical imaging applications, such annotated datasets are not easy to acquire, it takes a substantial amount of time and resource to curate an annotated medical image set. In this paper, we propose an efficient annotation framework for brain MR images that can suggest informative sample images for human experts to annotate. We evaluate the framework on two different brain image analysis tasks, namely brain tumour segmentation and whole brain segmentation. Experiments show that for brain tumour segmentation task on the BraTS 2019 dataset, training a segmentation model with only 7% suggestively annotated image samples can achieve a performance comparable to that of training on the full dataset. For whole brain segmentation on the MALC dataset, training with 42% suggestively annotated image samples can achieve a comparable performance to training on the full dataset. The proposed framework demonstrates a promising way to save manual annotation cost and improve data efficiency in medical imaging applications.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021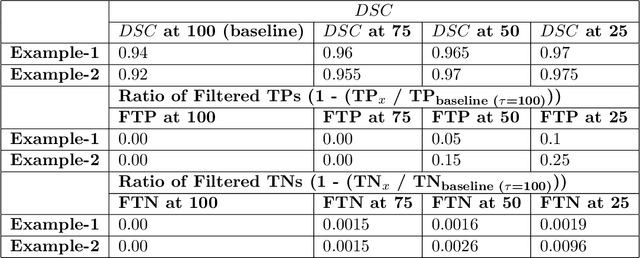
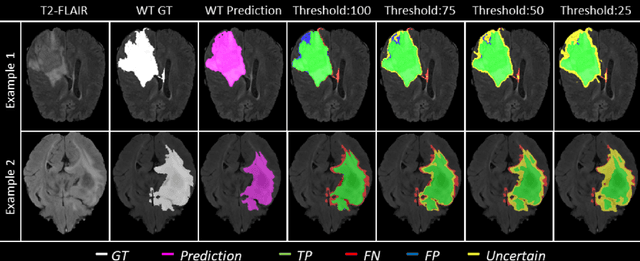
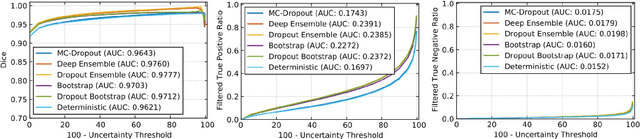
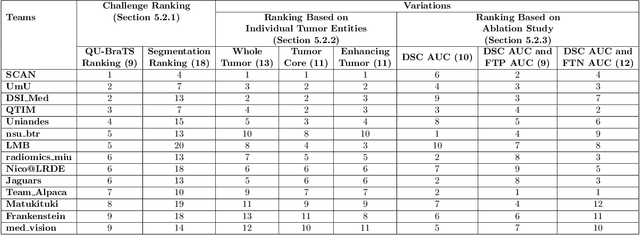
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Suggestive Annotation of Brain Tumour Images with Gradient-guided Sampling
Jul 03, 2020



Abstract:Machine learning has been widely adopted for medical image analysis in recent years given its promising performance in image segmentation and classification tasks. As a data-driven science, the success of machine learning, in particular supervised learning, largely depends on the availability of manually annotated datasets. For medical imaging applications, such annotated datasets are not easy to acquire. It takes a substantial amount of time and resource to curate an annotated medical image set. In this paper, we propose an efficient annotation framework for brain tumour images that is able to suggest informative sample images for human experts to annotate. Our experiments show that training a segmentation model with only 19% suggestively annotated patient scans from BraTS 2019 dataset can achieve a comparable performance to training a model on the full dataset for whole tumour segmentation task. It demonstrates a promising way to save manual annotation cost and improve data efficiency in medical imaging applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge