Elsa Angelini
TSI
SINETRA: a Versatile Framework for Evaluating Single Neuron Tracking in Behaving Animals
Nov 14, 2024Abstract:Accurately tracking neuronal activity in behaving animals presents significant challenges due to complex motions and background noise. The lack of annotated datasets limits the evaluation and improvement of such tracking algorithms. To address this, we developed SINETRA, a versatile simulator that generates synthetic tracking data for particles on a deformable background, closely mimicking live animal recordings. This simulator produces annotated 2D and 3D videos that reflect the intricate movements seen in behaving animals like Hydra Vulgaris. We evaluated four state-of-the-art tracking algorithms highlighting the current limitations of these methods in challenging scenarios and paving the way for improved cell tracking techniques in dynamic biological systems.
Curriculum Learning for Few-Shot Domain Adaptation in CT-based Airway Tree Segmentation
Nov 08, 2024



Abstract:Despite advances with deep learning (DL), automated airway segmentation from chest CT scans continues to face challenges in segmentation quality and generalization across cohorts. To address these, we propose integrating Curriculum Learning (CL) into airway segmentation networks, distributing the training set into batches according to ad-hoc complexity scores derived from CT scans and corresponding ground-truth tree features. We specifically investigate few-shot domain adaptation, targeting scenarios where manual annotation of a full fine-tuning dataset is prohibitively expensive. Results are reported on two large open-cohorts (ATM22 and AIIB23) with high performance using CL for full training (Source domain) and few-shot fine-tuning (Target domain), but with also some insights on potential detrimental effects if using a classic Bootstrapping scoring function or if not using proper scan sequencing.
Dense Self-Supervised Learning for Medical Image Segmentation
Jul 29, 2024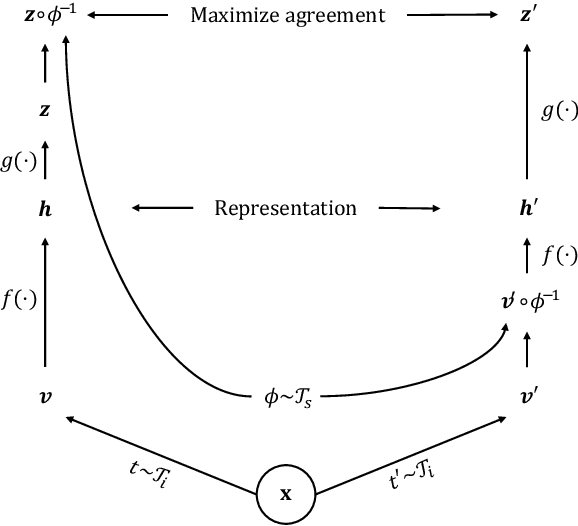

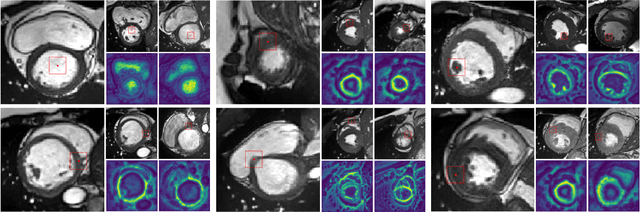

Abstract:Deep learning has revolutionized medical image segmentation, but it relies heavily on high-quality annotations. The time, cost and expertise required to label images at the pixel-level for each new task has slowed down widespread adoption of the paradigm. We propose Pix2Rep, a self-supervised learning (SSL) approach for few-shot segmentation, that reduces the manual annotation burden by learning powerful pixel-level representations directly from unlabeled images. Pix2Rep is a novel pixel-level loss and pre-training paradigm for contrastive SSL on whole images. It is applied to generic encoder-decoder deep learning backbones (e.g., U-Net). Whereas most SSL methods enforce invariance of the learned image-level representations under intensity and spatial image augmentations, Pix2Rep enforces equivariance of the pixel-level representations. We demonstrate the framework on a task of cardiac MRI segmentation. Results show improved performance compared to existing semi- and self-supervised approaches; and a 5-fold reduction in the annotation burden for equivalent performance versus a fully supervised U-Net baseline. This includes a 30% (resp. 31%) DICE improvement for one-shot segmentation under linear-probing (resp. fine-tuning). Finally, we also integrate the novel Pix2Rep concept with the Barlow Twins non-contrastive SSL, which leads to even better segmentation performance.
Deep ContourFlow: Advancing Active Contours with Deep Learning
Jul 15, 2024
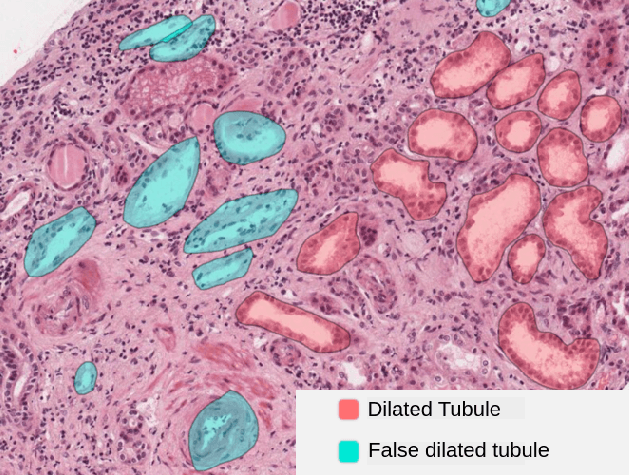

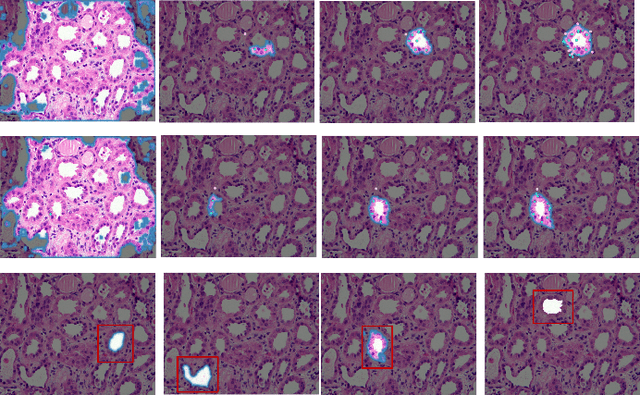
Abstract:This paper introduces a novel approach that combines unsupervised active contour models with deep learning for robust and adaptive image segmentation. Indeed, traditional active contours, provide a flexible framework for contour evolution and learning offers the capacity to learn intricate features and patterns directly from raw data. Our proposed methodology leverages the strengths of both paradigms, presenting a framework for both unsupervised and one-shot approaches for image segmentation. It is capable of capturing complex object boundaries without the need for extensive labeled training data. This is particularly required in histology, a field facing a significant shortage of annotations due to the challenging and time-consuming nature of the annotation process. We illustrate and compare our results to state of the art methods on a histology dataset and show significant improvements.
Few-Shot Airway-Tree Modeling using Data-Driven Sparse Priors
Jul 05, 2024



Abstract:The lack of large annotated datasets in medical imaging is an intrinsic burden for supervised Deep Learning (DL) segmentation models. Few-shot learning approaches are cost-effective solutions to transfer pre-trained models using only limited annotated data. However, such methods can be prone to overfitting due to limited data diversity especially when segmenting complex, diverse, and sparse tubular structures like airways. Furthermore, crafting informative image representations has played a crucial role in medical imaging, enabling discriminative enhancement of anatomical details. In this paper, we initially train a data-driven sparsification module to enhance airways efficiently in lung CT scans. We then incorporate these sparse representations in a standard supervised segmentation pipeline as a pretraining step to enhance the performance of the DL models. Results presented on the ATM public challenge cohort show the effectiveness of using sparse priors in pre-training, leading to segmentation Dice score increase by 1% to 10% in full-scale and few-shot learning scenarios, respectively.
Push the Boundary of SAM: A Pseudo-label Correction Framework for Medical Segmentation
Aug 02, 2023Abstract:Segment anything model (SAM) has emerged as the leading approach for zero-shot learning in segmentation, offering the advantage of avoiding pixel-wise annotation. It is particularly appealing in medical image segmentation where annotation is laborious and expertise-demanding. However, the direct application of SAM often yields inferior results compared to conventional fully supervised segmentation networks. While using SAM generated pseudo label could also benefit the training of fully supervised segmentation, the performance is limited by the quality of pseudo labels. In this paper, we propose a novel label corruption to push the boundary of SAM-based segmentation. Our model utilizes a novel noise detection module to distinguish between noisy labels from clean labels. This enables us to correct the noisy labels using an uncertainty-based self-correction module, thereby enriching the clean training set. Finally, we retrain the network with updated labels to optimize its weights for future predictions. One key advantage of our model is its ability to train deep networks using SAM-generated pseudo labels without relying on a subset of expert-level annotations. We demonstrate the effectiveness of our proposed model on both X-ray and lung CT datasets, indicating its ability to improve segmentation accuracy and outperform baseline methods in label correction.
Cardiac Adipose Tissue Segmentation via Image-Level Annotations
Jun 09, 2022



Abstract:Automatically identifying the structural substrates underlying cardiac abnormalities can potentially provide real-time guidance for interventional procedures. With the knowledge of cardiac tissue substrates, the treatment of complex arrhythmias such as atrial fibrillation and ventricular tachycardia can be further optimized by detecting arrhythmia substrates to target for treatment (i.e., adipose) and identifying critical structures to avoid. Optical coherence tomography (OCT) is a real-time imaging modality that aids in addressing this need. Existing approaches for cardiac image analysis mainly rely on fully supervised learning techniques, which suffer from the drawback of workload on labor-intensive annotation process of pixel-wise labeling. To lessen the need for pixel-wise labeling, we develop a two-stage deep learning framework for cardiac adipose tissue segmentation using image-level annotations on OCT images of human cardiac substrates. In particular, we integrate class activation mapping with superpixel segmentation to solve the sparse tissue seed challenge raised in cardiac tissue segmentation. Our study bridges the gap between the demand on automatic tissue analysis and the lack of high-quality pixel-wise annotations. To the best of our knowledge, this is the first study that attempts to address cardiac tissue segmentation on OCT images via weakly supervised learning techniques. Within an in-vitro human cardiac OCT dataset, we demonstrate that our weakly supervised approach on image-level annotations achieves comparable performance as fully supervised methods trained on pixel-wise annotations.
Suggestive Annotation of Brain MR Images with Gradient-guided Sampling
Jun 02, 2022



Abstract:Machine learning has been widely adopted for medical image analysis in recent years given its promising performance in image segmentation and classification tasks. The success of machine learning, in particular supervised learning, depends on the availability of manually annotated datasets. For medical imaging applications, such annotated datasets are not easy to acquire, it takes a substantial amount of time and resource to curate an annotated medical image set. In this paper, we propose an efficient annotation framework for brain MR images that can suggest informative sample images for human experts to annotate. We evaluate the framework on two different brain image analysis tasks, namely brain tumour segmentation and whole brain segmentation. Experiments show that for brain tumour segmentation task on the BraTS 2019 dataset, training a segmentation model with only 7% suggestively annotated image samples can achieve a performance comparable to that of training on the full dataset. For whole brain segmentation on the MALC dataset, training with 42% suggestively annotated image samples can achieve a comparable performance to training on the full dataset. The proposed framework demonstrates a promising way to save manual annotation cost and improve data efficiency in medical imaging applications.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021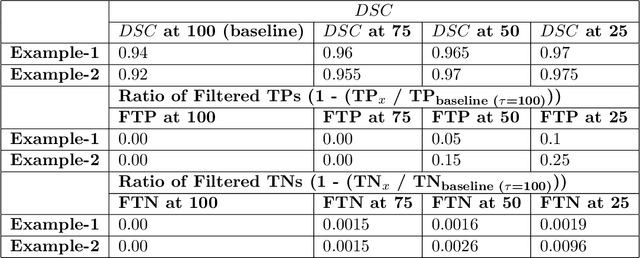
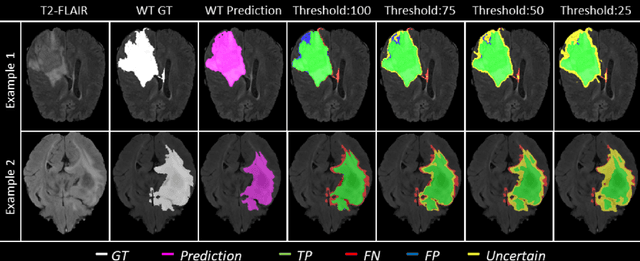
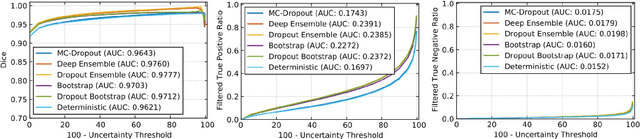
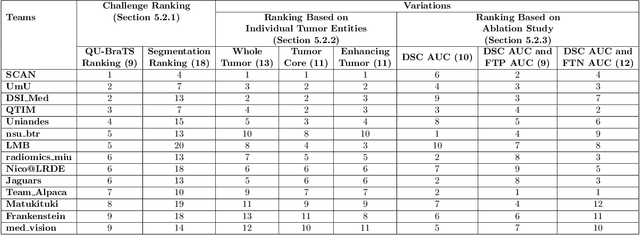
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Recursive Refinement Network for Deformable Lung Registration between Exhale and Inhale CT Scans
Jun 14, 2021
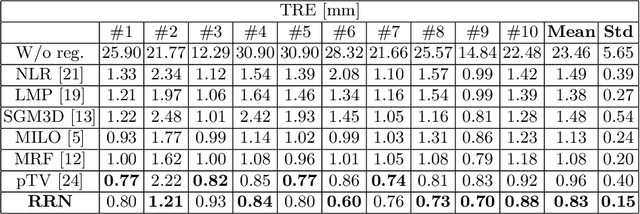
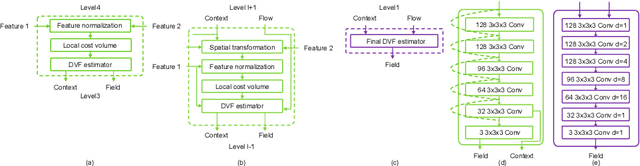
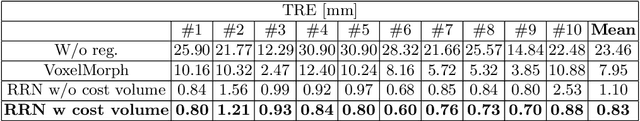
Abstract:Unsupervised learning-based medical image registration approaches have witnessed rapid development in recent years. We propose to revisit a commonly ignored while simple and well-established principle: recursive refinement of deformation vector fields across scales. We introduce a recursive refinement network (RRN) for unsupervised medical image registration, to extract multi-scale features, construct normalized local cost correlation volume and recursively refine volumetric deformation vector fields. RRN achieves state of the art performance for 3D registration of expiratory-inspiratory pairs of CT lung scans. On DirLab COPDGene dataset, RRN returns an average Target Registration Error (TRE) of 0.83 mm, which corresponds to a 13% error reduction from the best result presented in the leaderboard. In addition to comparison with conventional methods, RRN leads to 89% error reduction compared to deep-learning-based peer approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge