Maxime Seince
Discriminative protein sequence modelling with Latent Space Diffusion
Mar 24, 2025



Abstract:We explore a framework for protein sequence representation learning that decomposes the task between manifold learning and distributional modelling. Specifically we present a Latent Space Diffusion architecture which combines a protein sequence autoencoder with a denoising diffusion model operating on its latent space. We obtain a one-parameter family of learned representations from the diffusion model, along with the autoencoder's latent representation. We propose and evaluate two autoencoder architectures: a homogeneous model forcing amino acids of the same type to be identically distributed in the latent space, and an inhomogeneous model employing a noise-based variant of masking. As a baseline we take a latent space learned by masked language modelling, and evaluate discriminative capability on a range of protein property prediction tasks. Our finding is twofold: the diffusion models trained on both our proposed variants display higher discriminative power than the one trained on the masked language model baseline, none of the diffusion representations achieve the performance of the masked language model embeddings themselves.
Dense Self-Supervised Learning for Medical Image Segmentation
Jul 29, 2024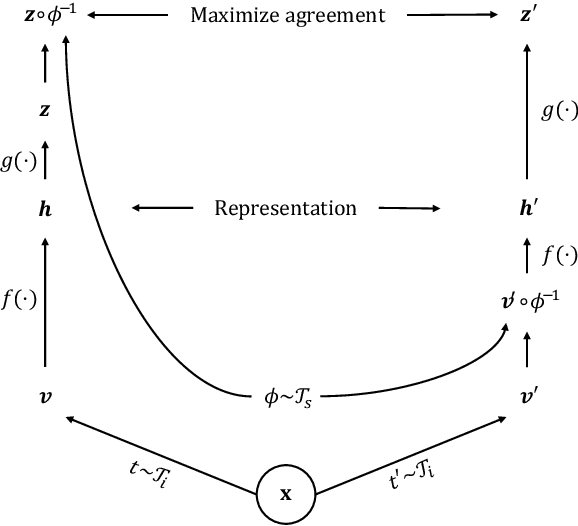

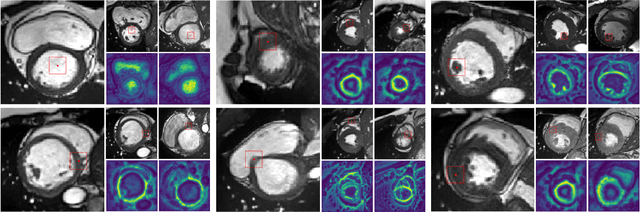

Abstract:Deep learning has revolutionized medical image segmentation, but it relies heavily on high-quality annotations. The time, cost and expertise required to label images at the pixel-level for each new task has slowed down widespread adoption of the paradigm. We propose Pix2Rep, a self-supervised learning (SSL) approach for few-shot segmentation, that reduces the manual annotation burden by learning powerful pixel-level representations directly from unlabeled images. Pix2Rep is a novel pixel-level loss and pre-training paradigm for contrastive SSL on whole images. It is applied to generic encoder-decoder deep learning backbones (e.g., U-Net). Whereas most SSL methods enforce invariance of the learned image-level representations under intensity and spatial image augmentations, Pix2Rep enforces equivariance of the pixel-level representations. We demonstrate the framework on a task of cardiac MRI segmentation. Results show improved performance compared to existing semi- and self-supervised approaches; and a 5-fold reduction in the annotation burden for equivalent performance versus a fully supervised U-Net baseline. This includes a 30% (resp. 31%) DICE improvement for one-shot segmentation under linear-probing (resp. fine-tuning). Finally, we also integrate the novel Pix2Rep concept with the Barlow Twins non-contrastive SSL, which leads to even better segmentation performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge