Fuyong Xing
Push the Boundary of SAM: A Pseudo-label Correction Framework for Medical Segmentation
Aug 02, 2023Abstract:Segment anything model (SAM) has emerged as the leading approach for zero-shot learning in segmentation, offering the advantage of avoiding pixel-wise annotation. It is particularly appealing in medical image segmentation where annotation is laborious and expertise-demanding. However, the direct application of SAM often yields inferior results compared to conventional fully supervised segmentation networks. While using SAM generated pseudo label could also benefit the training of fully supervised segmentation, the performance is limited by the quality of pseudo labels. In this paper, we propose a novel label corruption to push the boundary of SAM-based segmentation. Our model utilizes a novel noise detection module to distinguish between noisy labels from clean labels. This enables us to correct the noisy labels using an uncertainty-based self-correction module, thereby enriching the clean training set. Finally, we retrain the network with updated labels to optimize its weights for future predictions. One key advantage of our model is its ability to train deep networks using SAM-generated pseudo labels without relying on a subset of expert-level annotations. We demonstrate the effectiveness of our proposed model on both X-ray and lung CT datasets, indicating its ability to improve segmentation accuracy and outperform baseline methods in label correction.
Self-paced Resistance Learning against Overfitting on Noisy Labels
May 07, 2021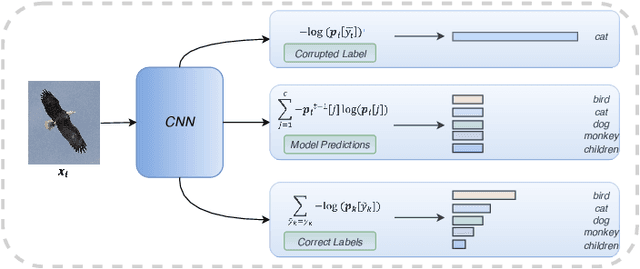
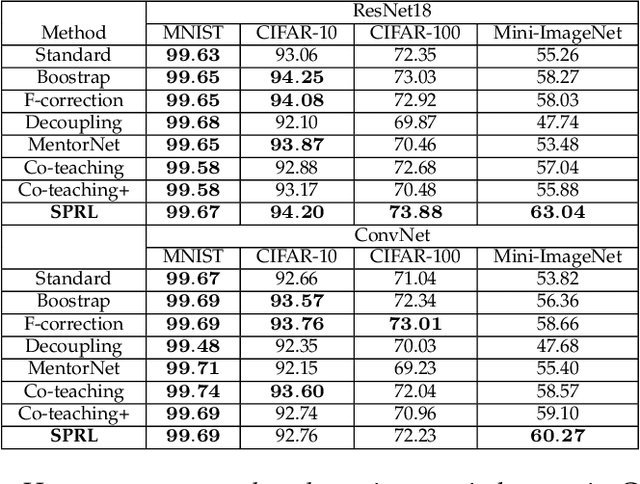
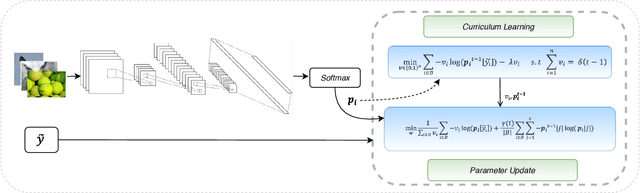
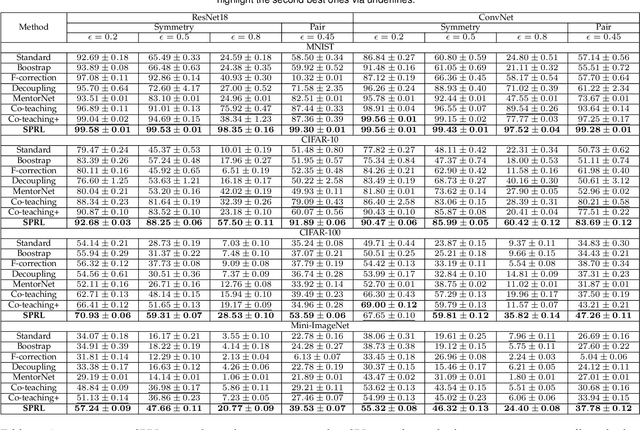
Abstract:Noisy labels composed of correct and corrupted ones are pervasive in practice. They might significantly deteriorate the performance of convolutional neural networks (CNNs), because CNNs are easily overfitted on corrupted labels. To address this issue, inspired by an observation, deep neural networks might first memorize the probably correct-label data and then corrupt-label samples, we propose a novel yet simple self-paced resistance framework to resist corrupted labels, without using any clean validation data. The proposed framework first utilizes the memorization effect of CNNs to learn a curriculum, which contains confident samples and provides meaningful supervision for other training samples. Then it adopts selected confident samples and a proposed resistance loss to update model parameters; the resistance loss tends to smooth model parameters' update or attain equivalent prediction over each class, thereby resisting model overfitting on corrupted labels. Finally, we unify these two modules into a single loss function and optimize it in an alternative learning. Extensive experiments demonstrate the significantly superior performance of the proposed framework over recent state-of-the-art methods on noisy-label data. Source codes of the proposed method are available on https://github.com/xsshi2015/Self-paced-Resistance-Learning.
Cluster Activation Mapping with Applications to Medical Imaging
Oct 09, 2020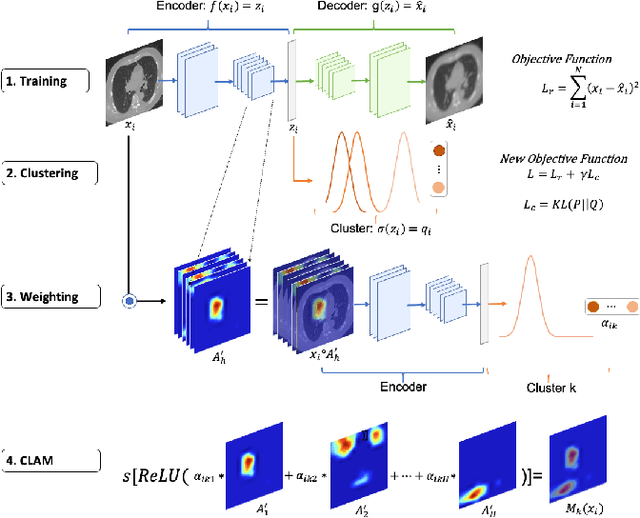
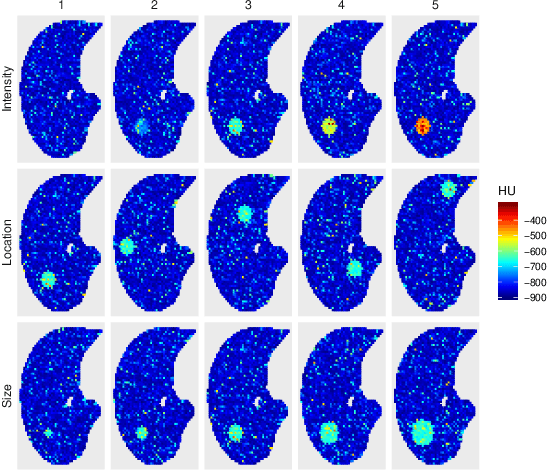
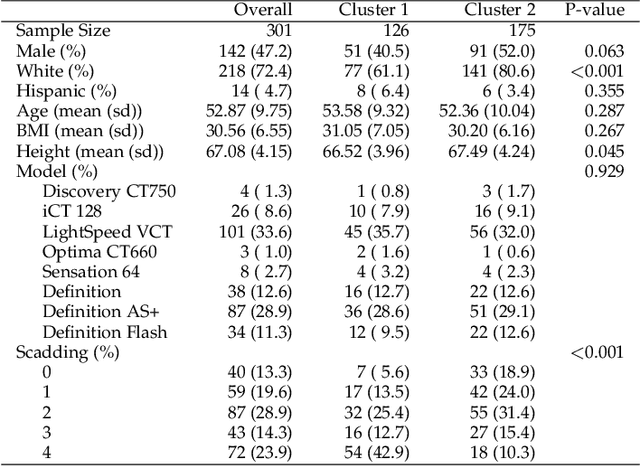
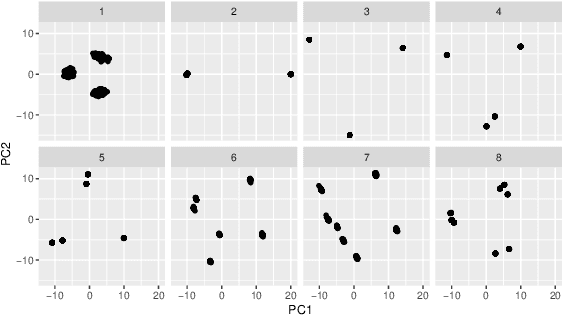
Abstract:An open question in deep clustering is how to understand what in the image is creating the cluster assignments. This visual understanding is essential to be able to trust the results of an inherently complex algorithm like deep learning, especially when the derived cluster assignments may be used to inform decision-making or create new disease sub-types. In this work, we developed novel methodology to generate CLuster Activation Mapping (CLAM) which combines an unsupervised deep clustering framework with a modification of Score-CAM, an approach for discriminative localization in the supervised setting. We evaluated our approach using a simulation study based on computed tomography scans of the lung, and applied it to 3D CT scans from a sarcoidosis population to identify new clusters of sarcoidosis based purely on CT scan presentation.
Pancreas Segmentation in CT and MRI Images via Domain Specific Network Designing and Recurrent Neural Contextual Learning
Mar 30, 2018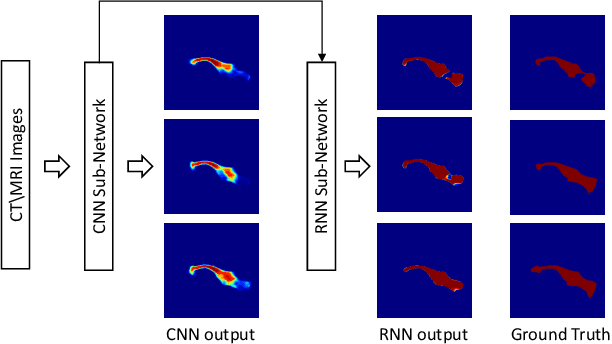
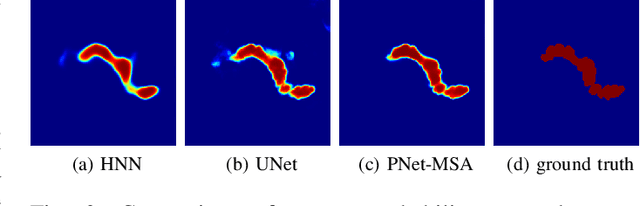
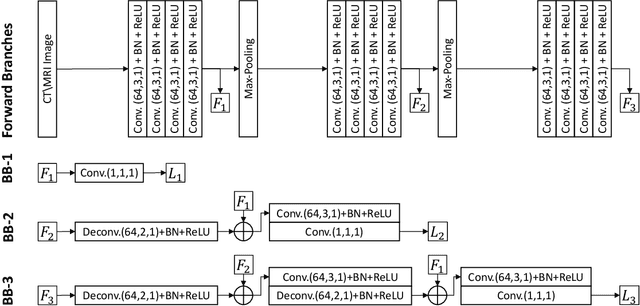
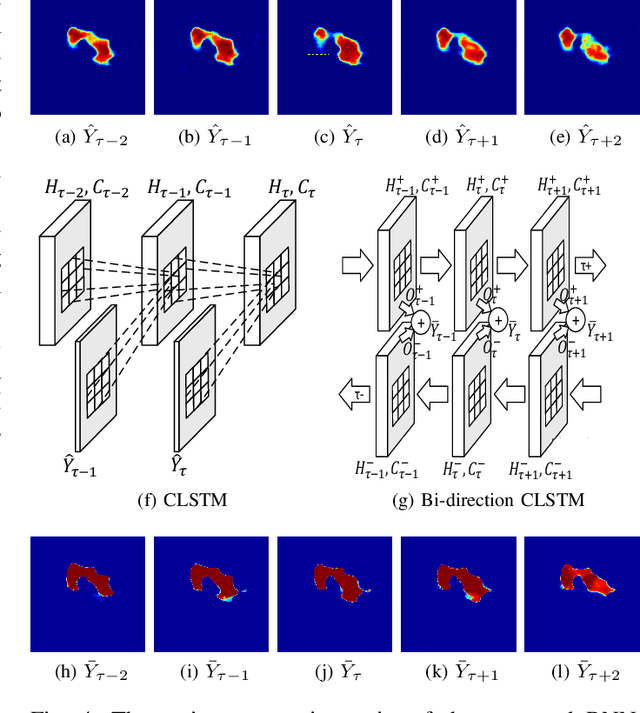
Abstract:Automatic pancreas segmentation in radiology images, eg., computed tomography (CT) and magnetic resonance imaging (MRI), is frequently required by computer-aided screening, diagnosis, and quantitative assessment. Yet pancreas is a challenging abdominal organ to segment due to the high inter-patient anatomical variability in both shape and volume metrics. Recently, convolutional neural networks (CNNs) have demonstrated promising performance on accurate segmentation of pancreas. However, the CNN-based method often suffers from segmentation discontinuity for reasons such as noisy image quality and blurry pancreatic boundary. From this point, we propose to introduce recurrent neural networks (RNNs) to address the problem of spatial non-smoothness of inter-slice pancreas segmentation across adjacent image slices. To inference initial segmentation, we first train a 2D CNN sub-network, where we modify its network architecture with deep-supervision and multi-scale feature map aggregation so that it can be trained from scratch with small-sized training data and presents superior performance than transferred models. Thereafter, the successive CNN outputs are processed by another RNN sub-network, which refines the consistency of segmented shapes. More specifically, the RNN sub-network consists convolutional long short-term memory (CLSTM) units in both top-down and bottom-up directions, which regularizes the segmentation of an image by integrating predictions of its neighboring slices. We train the stacked CNN-RNN model end-to-end and perform quantitative evaluations on both CT and MRI images.
Revisiting Graph Construction for Fast Image Segmentation
Dec 02, 2017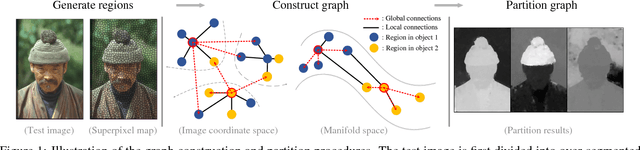
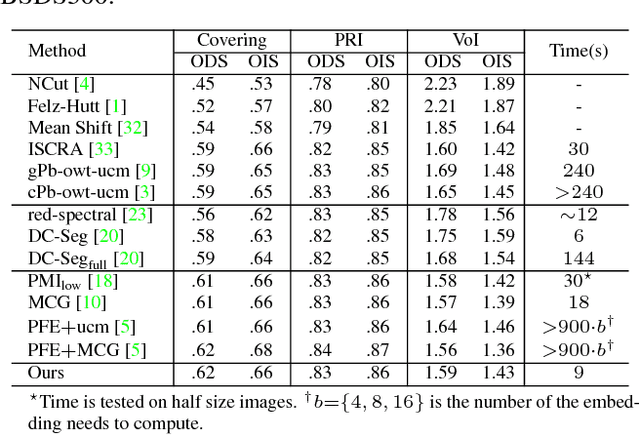
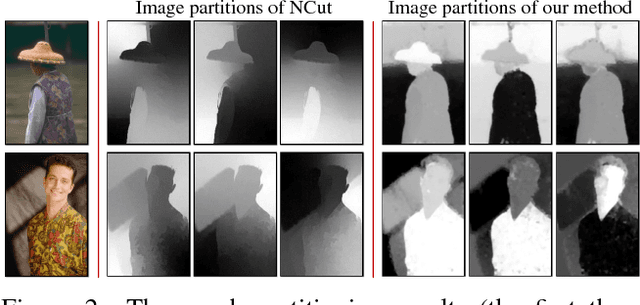
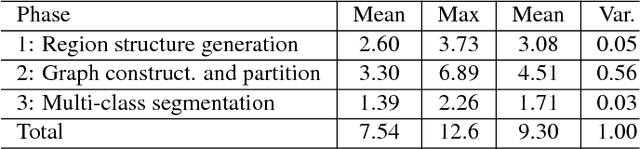
Abstract:In this paper, we propose a simple but effective method for fast image segmentation. We re-examine the locality-preserving character of spectral clustering by constructing a graph over image regions with both global and local connections. Our novel approach to build graph connections relies on two key observations: 1) local region pairs that co-occur frequently will have a high probability to reside on a common object; 2) spatially distant regions in a common object often exhibit similar visual saliency, which implies their neighborship in a manifold. We present a novel energy function to efficiently conduct graph partitioning. Based on multiple high quality partitions, we show that the generated eigenvector histogram based representation can automatically drive effective unary potentials for a hierarchical random field model to produce multi-class segmentation. Sufficient experiments, on the BSDS500 benchmark, large-scale PASCAL VOC and COCO datasets, demonstrate the competitive segmentation accuracy and significantly improved efficiency of our proposed method compared with other state of the arts.
Recent Advances in the Applications of Convolutional Neural Networks to Medical Image Contour Detection
Aug 24, 2017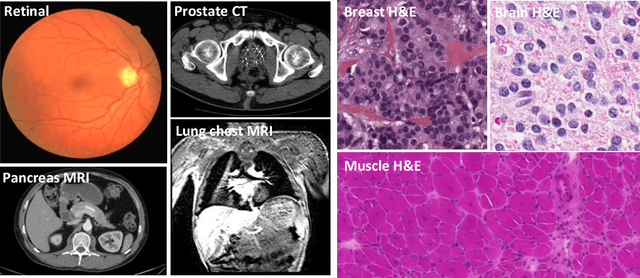
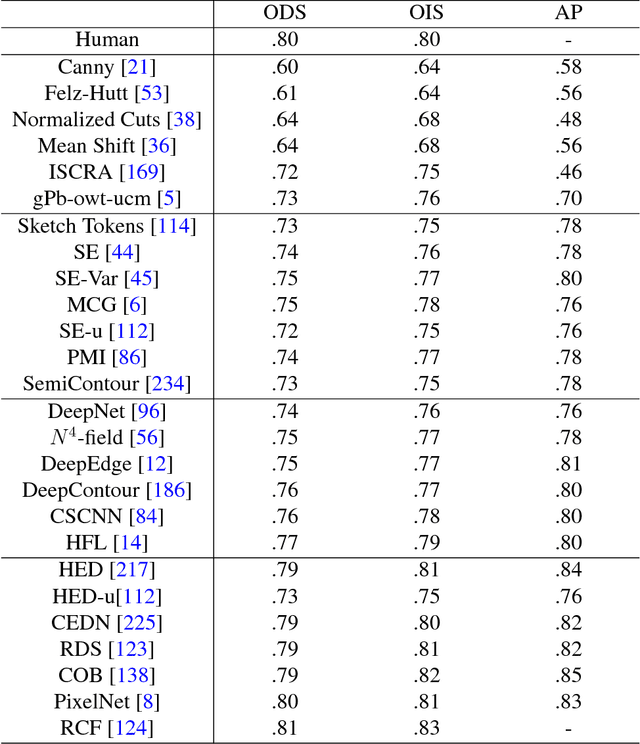
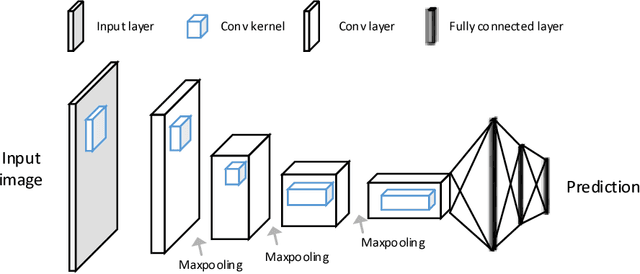
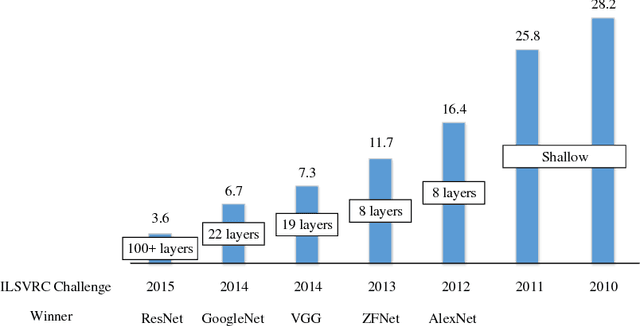
Abstract:The fast growing deep learning technologies have become the main solution of many machine learning problems for medical image analysis. Deep convolution neural networks (CNNs), as one of the most important branch of the deep learning family, have been widely investigated for various computer-aided diagnosis tasks including long-term problems and continuously emerging new problems. Image contour detection is a fundamental but challenging task that has been studied for more than four decades. Recently, we have witnessed the significantly improved performance of contour detection thanks to the development of CNNs. Beyond purusing performance in existing natural image benchmarks, contour detection plays a particularly important role in medical image analysis. Segmenting various objects from radiology images or pathology images requires accurate detection of contours. However, some problems, such as discontinuity and shape constraints, are insufficiently studied in CNNs. It is necessary to clarify the challenges to encourage further exploration. The performance of CNN based contour detection relies on the state-of-the-art CNN architectures. Careful investigation of their design principles and motivations is critical and beneficial to contour detection. In this paper, we first review recent development of medical image contour detection and point out the current confronting challenges and problems. We discuss the development of general CNNs and their applications in image contours (or edges) detection. We compare those methods in detail, clarify their strengthens and weaknesses. Then we review their recent applications in medical image analysis and point out limitations, with the goal to light some potential directions in medical image analysis. We expect the paper to cover comprehensive technical ingredients of advanced CNNs to enrich the study in the medical image domain.
Improving Deep Pancreas Segmentation in CT and MRI Images via Recurrent Neural Contextual Learning and Direct Loss Function
Jul 18, 2017


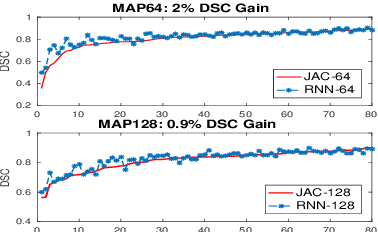
Abstract:Deep neural networks have demonstrated very promising performance on accurate segmentation of challenging organs (e.g., pancreas) in abdominal CT and MRI scans. The current deep learning approaches conduct pancreas segmentation by processing sequences of 2D image slices independently through deep, dense per-pixel masking for each image, without explicitly enforcing spatial consistency constraint on segmentation of successive slices. We propose a new convolutional/recurrent neural network architecture to address the contextual learning and segmentation consistency problem. A deep convolutional sub-network is first designed and pre-trained from scratch. The output layer of this network module is then connected to recurrent layers and can be fine-tuned for contextual learning, in an end-to-end manner. Our recurrent sub-network is a type of Long short-term memory (LSTM) network that performs segmentation on an image by integrating its neighboring slice segmentation predictions, in the form of a dependent sequence processing. Additionally, a novel segmentation-direct loss function (named Jaccard Loss) is proposed and deep networks are trained to optimize Jaccard Index (JI) directly. Extensive experiments are conducted to validate our proposed deep models, on quantitative pancreas segmentation using both CT and MRI scans. Our method outperforms the state-of-the-art work on CT [11] and MRI pancreas segmentation [1], respectively.
MDNet: A Semantically and Visually Interpretable Medical Image Diagnosis Network
Jul 08, 2017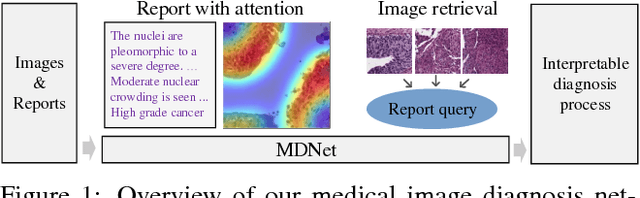
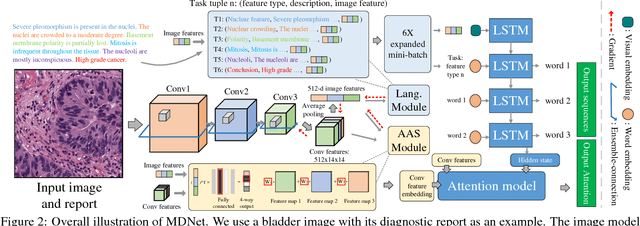
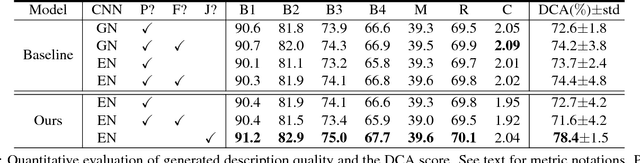

Abstract:The inability to interpret the model prediction in semantically and visually meaningful ways is a well-known shortcoming of most existing computer-aided diagnosis methods. In this paper, we propose MDNet to establish a direct multimodal mapping between medical images and diagnostic reports that can read images, generate diagnostic reports, retrieve images by symptom descriptions, and visualize attention, to provide justifications of the network diagnosis process. MDNet includes an image model and a language model. The image model is proposed to enhance multi-scale feature ensembles and utilization efficiency. The language model, integrated with our improved attention mechanism, aims to read and explore discriminative image feature descriptions from reports to learn a direct mapping from sentence words to image pixels. The overall network is trained end-to-end by using our developed optimization strategy. Based on a pathology bladder cancer images and its diagnostic reports (BCIDR) dataset, we conduct sufficient experiments to demonstrate that MDNet outperforms comparative baselines. The proposed image model obtains state-of-the-art performance on two CIFAR datasets as well.
SemiContour: A Semi-supervised Learning Approach for Contour Detection
May 17, 2016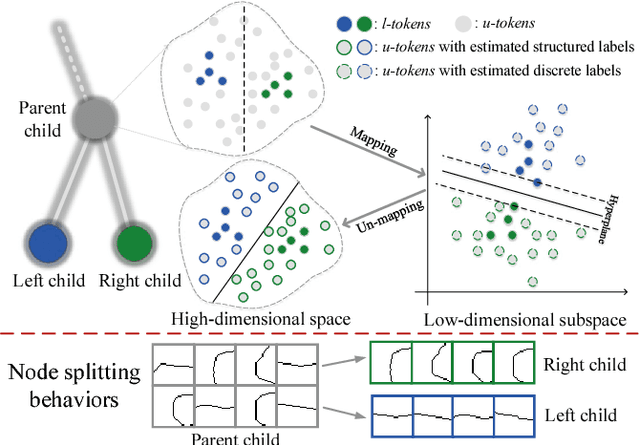
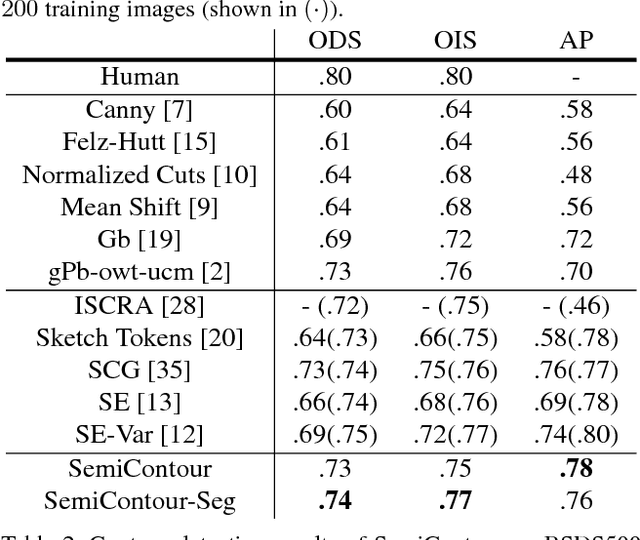

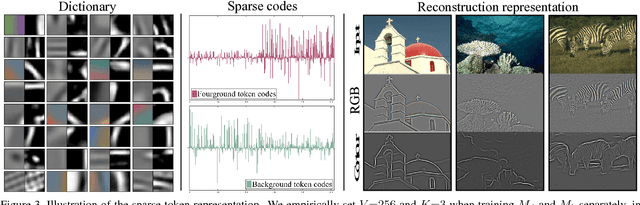
Abstract:Supervised contour detection methods usually require many labeled training images to obtain satisfactory performance. However, a large set of annotated data might be unavailable or extremely labor intensive. In this paper, we investigate the usage of semi-supervised learning (SSL) to obtain competitive detection accuracy with very limited training data (three labeled images). Specifically, we propose a semi-supervised structured ensemble learning approach for contour detection built on structured random forests (SRF). To allow SRF to be applicable to unlabeled data, we present an effective sparse representation approach to capture inherent structure in image patches by finding a compact and discriminative low-dimensional subspace representation in an unsupervised manner, enabling the incorporation of abundant unlabeled patches with their estimated structured labels to help SRF perform better node splitting. We re-examine the role of sparsity and propose a novel and fast sparse coding algorithm to boost the overall learning efficiency. To the best of our knowledge, this is the first attempt to apply SSL for contour detection. Extensive experiments on the BSDS500 segmentation dataset and the NYU Depth dataset demonstrate the superiority of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge