Pancreas Segmentation in CT and MRI Images via Domain Specific Network Designing and Recurrent Neural Contextual Learning
Paper and Code
Mar 30, 2018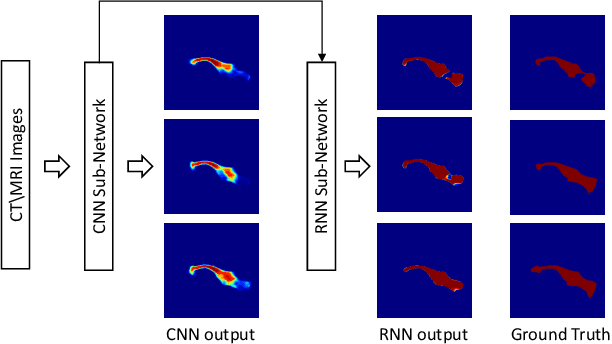
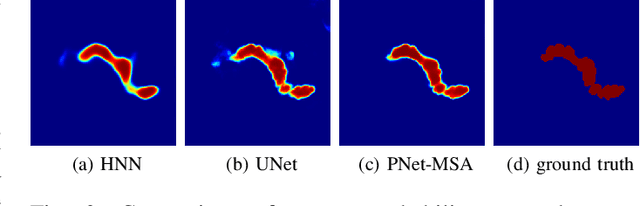
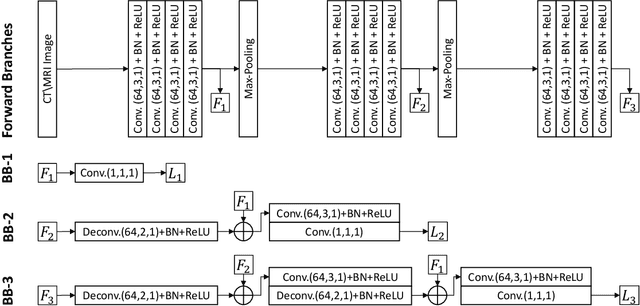
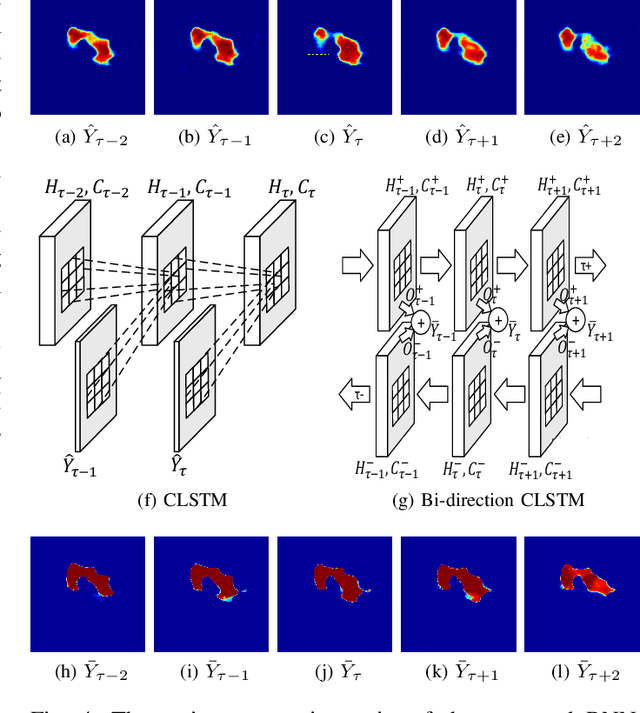
Automatic pancreas segmentation in radiology images, eg., computed tomography (CT) and magnetic resonance imaging (MRI), is frequently required by computer-aided screening, diagnosis, and quantitative assessment. Yet pancreas is a challenging abdominal organ to segment due to the high inter-patient anatomical variability in both shape and volume metrics. Recently, convolutional neural networks (CNNs) have demonstrated promising performance on accurate segmentation of pancreas. However, the CNN-based method often suffers from segmentation discontinuity for reasons such as noisy image quality and blurry pancreatic boundary. From this point, we propose to introduce recurrent neural networks (RNNs) to address the problem of spatial non-smoothness of inter-slice pancreas segmentation across adjacent image slices. To inference initial segmentation, we first train a 2D CNN sub-network, where we modify its network architecture with deep-supervision and multi-scale feature map aggregation so that it can be trained from scratch with small-sized training data and presents superior performance than transferred models. Thereafter, the successive CNN outputs are processed by another RNN sub-network, which refines the consistency of segmented shapes. More specifically, the RNN sub-network consists convolutional long short-term memory (CLSTM) units in both top-down and bottom-up directions, which regularizes the segmentation of an image by integrating predictions of its neighboring slices. We train the stacked CNN-RNN model end-to-end and perform quantitative evaluations on both CT and MRI images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge